| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
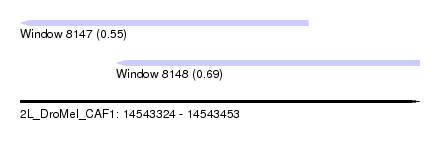
| Location | 14,543,324 – 14,543,453 |
| Length | 129 |
| Max. P | 0.687815 |

| Location | 14,543,324 – 14,543,417 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 69.65 |
| Mean single sequence MFE | -24.32 |
| Consensus MFE | -6.98 |
| Energy contribution | -8.40 |
| Covariance contribution | 1.42 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.29 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.548008 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
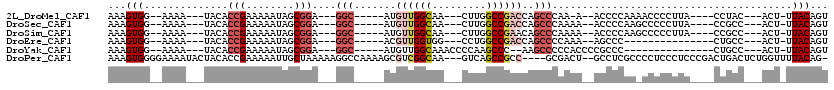
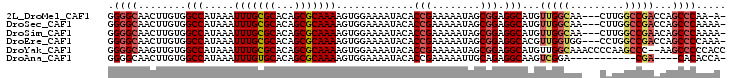
>2L_DroMel_CAF1 14543324 93 - 22407834 AAAGUGG--AAAA---UACACCGAAAAAUAGCGGA---GGC-----AUGUUGGCAA---CUUGGCCGACCAGCCCAA-A--ACCCCAAAACCCCUUA----CCUAC---ACU-UUACAGU ((((((.--....---....(((........))).---(((-----..((((((..---....))))))..)))...-.--................----....)---)))-))..... ( -22.00) >DroSec_CAF1 75194 94 - 1 AAAGUGG--AAAA---UACACCGAAAAAUAGCGGA---GGC-----AUGUUGGCAA---CUUGGCCGACCAGCCCAAAA--ACCCCAAGCCCCCUUA----CCGCC---ACU-UUACAGU (((((((--....---....(((........))).---(((-----..((((((..---....))))))..))).....--................----...))---)))-))..... ( -24.80) >DroSim_CAF1 61011 94 - 1 AAAGUGG--AAAA---UACACCGAAAAAUAGCGGA---GGC-----AUGUUGGCAA---CUUGGCCGAACAGCCCAAAA--ACCCCAAGCCCCCUUA----CCGCC---ACU-UUACAGU (((((((--....---....(((........))).---(((-----.(((((((..---....))).))))))).....--................----...))---)))-))..... ( -23.60) >DroEre_CAF1 70232 83 - 1 AAAGUGG--AAAA---UACACCGAAAAAUAGCGGA---GGC-----ACGUUGGUGG---CCUGGCCGACCAGCCCCAAA--AGCCC---------------CUGCC---ACU-UUACAGU (((((((--....---....(((........))).---(((-----...((((.((---(.(((....)))))))))).--.))).---------------...))---)))-))..... ( -28.90) >DroYak_CAF1 66134 86 - 1 AAAGUGG--AAAA---UACACCGAAAAAUAGCGGA---GGC-----AUGUUGGCAAACCCCAAGCCC--AAGCCCCCACCCCGCCC---------------CUGCC---ACU-UUACAGU (((((((--....---....(((........))).---(((-----.((..(((.........))))--).)))............---------------...))---)))-))..... ( -19.10) >DroPer_CAF1 87761 110 - 1 AAAGUGGGGAAAAUACUACACCGAAAAAUUGCUAAAAAGGCCAAAAGCGUCGGCAA---GUCAGCCGCC----GCGACU--GCCUCGCCCCUCCCUCCCGACUGACUCUGGUUUUACAG- ..(((.((((...........(((....)))......((((.....(((.((((..---....)))).)----))....--))))..........)))).)))....(((......)))- ( -27.50) >consensus AAAGUGG__AAAA___UACACCGAAAAAUAGCGGA___GGC_____AUGUUGGCAA___CUUGGCCGACCAGCCCAAAA__ACCCCAA_CCCCCUUA____CCGCC___ACU_UUACAGU ...(((..............(((........)))....(((.......((((((.........))))))..)))........................................)))... ( -6.98 = -8.40 + 1.42)
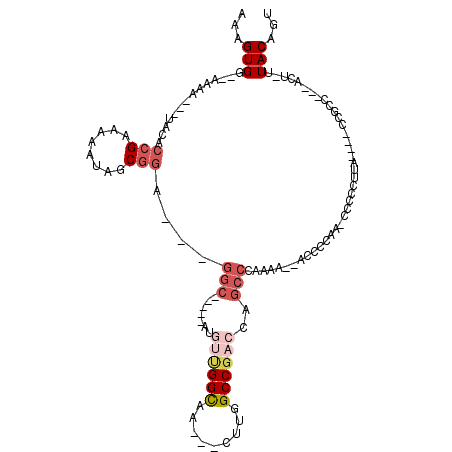
| Location | 14,543,355 – 14,543,453 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 86.70 |
| Mean single sequence MFE | -32.88 |
| Consensus MFE | -23.94 |
| Energy contribution | -24.75 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.14 |
| Mean z-score | -3.16 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.687815 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14543355 98 - 22407834 GGGGCAACUUGUGGCCAUAAAUUUGCGCACAGCGCAAAAGUGGAAAAUACACCGAAAAAUAGCGGAGGCAUGUUGGCAA---CUUGGCCGACCAGCCCAA-A- .((((........(((.....(((((((...))))))).............(((........))).)))..((((((..---....))))))..))))..-.- ( -34.10) >DroSec_CAF1 75225 99 - 1 GGGGCAACUUGUGGCCAUAAAUUUGCGCACAGCGCAAAAGUGGAAAAUACACCGAAAAAUAGCGGAGGCAUGUUGGCAA---CUUGGCCGACCAGCCCAAAA- .((((........(((.....(((((((...))))))).............(((........))).)))..((((((..---....))))))..))))....- ( -34.10) >DroSim_CAF1 61042 99 - 1 GGGGCAACUUGUGGCCAUAAAUUUGCGCACAGCGCAAAAGUGGAAAAUACACCGAAAAAUAGCGGAGGCAUGUUGGCAA---CUUGGCCGAACAGCCCAAAA- .((((........(((.....(((((((...))))))).............(((........))).))).(((((((..---....))).))))))))....- ( -32.50) >DroEre_CAF1 70252 99 - 1 GGGGCAACUUGUGGCCAUAAAUUUGCGCACAGCGCAAAAGUGGAAAAUACACCGAAAAAUAGCGGAGGCACGUUGGUGG---CCUGGCCGACCAGCCCCAAA- (((((.......((((((...(((((((...))))))).(((.......))).......(((((......)))))))))---))(((....))))))))...- ( -37.80) >DroYak_CAF1 66155 101 - 1 GGGGCAAGUUGUGGCCAUAAAUUUGCGCACAGCGCAAAAGUGGAAAAUACACCGAAAAAUAGCGGAGGCAUGUUGGCAAACCCCAAGCCC--AAGCCCCCACC (((((....((((.((((...(((((((...))))))).))))....))))(((........))).(((...((((......))))))).--..))))).... ( -33.40) >DroAna_CAF1 13784 87 - 1 GGGGCAACUUGUGGCCAUAAAUUUGUGCACAGCGCAAAAGUGGAAAAUACACCGAAAAAUUGCAGAGGCAAGUCGGA-----------CGA----CACACCA- (((....).((((.((((...(((((((...))))))).))))........((((....((((....)))).)))).-----------...----)))))).- ( -25.40) >consensus GGGGCAACUUGUGGCCAUAAAUUUGCGCACAGCGCAAAAGUGGAAAAUACACCGAAAAAUAGCGGAGGCAUGUUGGCAA___CUUGGCCGACCAGCCCAAAA_ .((((........(((.....(((((((...))))))).............(((........))).)))...(((((.........)))))...))))..... (-23.94 = -24.75 + 0.81)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:36:26 2006