| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,541,672 – 14,541,782 |
| Length | 110 |
| Max. P | 0.773254 |

| Location | 14,541,672 – 14,541,782 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 113 |
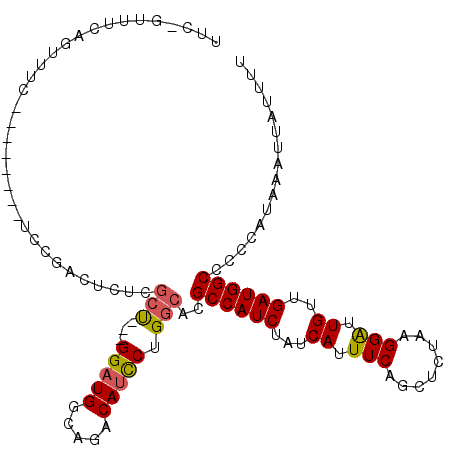
| Reading direction | reverse |
| Mean pairwise identity | 78.40 |
| Mean single sequence MFE | -25.59 |
| Consensus MFE | -18.99 |
| Energy contribution | -18.88 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.773254 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14541672 110 - 22407834 UUCGGUUUCAGUUUCAGUUUUAGUCCGACUCUCGCU---GGAUGGCAGACAUCCUGGCACGCCAUCUAUCAUUUCAGCUCUAAGGAUUGUUGAUGGCCCCCAUAAAUUAUUUU .((((...................)))).....(((---(((((.....))))).)))..((((((...((.(((........))).))..))))))................ ( -25.51) >DroPse_CAF1 80943 91 - 1 C----------------------UCUGACUCUGGCUCUGGGAUGGCAGACAUUCCGGCACGCCAUCUAUCAUUUCAGCUCUAAGGGUUGUUGAUGGCCUCCAUAAAUUAUAUU .----------------------........((((.((((((((.....))))))))...)))).((((((...(((((.....))))).))))))................. ( -26.50) >DroEre_CAF1 68592 110 - 1 UUCAGUUUCAGUUUCAGUUUGGGUCCGACUCUCGCU---GGCUGGCAGACAUCCUGGCACGCCAUCUAUCAUUUCAGCUCUAAGGAUUGUUGAUGGCACCCAUAAAUUAUUUU ....((((((((..((((..(((.....)))..)))---))))))..)))....(((...((((((...((.(((........))).))..))))))..)))........... ( -26.20) >DroYak_CAF1 64425 110 - 1 UUCGGUUUCGGUUUCAGUUGUAGUCCAACUCUCGCU---GGAUGGCAGACAUCCUGGCACGCCAUCUAUCAUUUCAGCUCUAAGGAUUGUUGAUGGCCCCCAUAAAUUAUUUU .........((....(((((.....)))))...(((---(((((.....))))).)))..((((((...((.(((........))).))..))))))..))............ ( -26.70) >DroAna_CAF1 11974 93 - 1 UUCAGUUUCAGUCUC--------------UUU---C---GGAUGGCAGACAUCCUGGCACGCCAUCUAUCAUUUCAGCUCUAAGGGUUGUUGAUGGCCCCCAUAAAUUAUUUU ...............--------------...---.---(((((.....)))))(((...((((((........(((((.....)))))..))))))..)))........... ( -22.10) >DroPer_CAF1 85333 91 - 1 C----------------------UCUGACUCUGGCUCUGGGAUGGCAGACAUUCCGGCACGCCAUCUAUCAUUUCAGCUCUAAGGGUUGUUGAUGGCCUCCAUAAAUUAUAUU .----------------------........((((.((((((((.....))))))))...)))).((((((...(((((.....))))).))))))................. ( -26.50) >consensus UUC_GUUUCAGUUUC________UCCGACUCUCGCU___GGAUGGCAGACAUCCUGGCACGCCAUCUAUCAUUUCAGCUCUAAGGAUUGUUGAUGGCCCCCAUAAAUUAUUUU .................................(((...(((((.....))))).)))..((((((...((.(((........))).))..))))))................ (-18.99 = -18.88 + -0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:36:23 2006