
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,535,787 – 14,535,918 |
| Length | 131 |
| Max. P | 0.950241 |

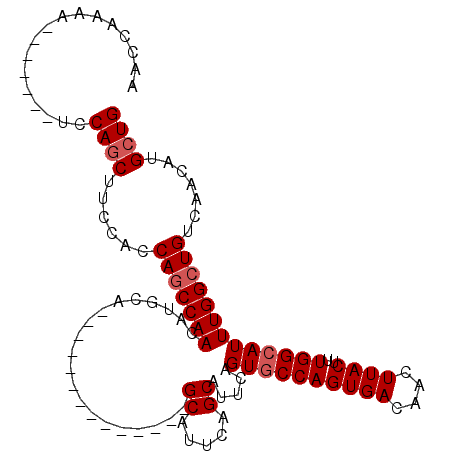
| Location | 14,535,787 – 14,535,878 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 85.06 |
| Mean single sequence MFE | -22.89 |
| Consensus MFE | -18.84 |
| Energy contribution | -19.59 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.950241 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
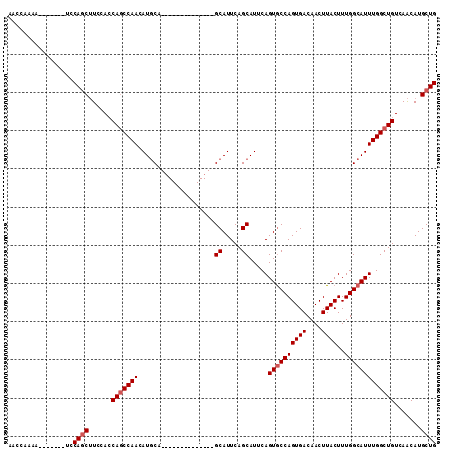
>2L_DroMel_CAF1 14535787 91 + 22407834 AGCCAAAA-------UCCAGCUUCCACCAGCCAACAUUCA--------------GCAUUCAGCAUUCAGUGCCAGUGACAACUUACUUUGGCAUUUGGCUGUCAACAUGCUG ........-------..((((......(((((((......--------------((.....)).....((((((((((....))))..))))))))))))).......)))) ( -24.02) >DroSec_CAF1 67819 91 + 1 AACCGAAA-------UCCAGCUUCCACCAGCCAACAUGCA--------------GCAUUCAGCAUUCAGUGCCAGUGACAACUUACUUUGGCAUUUGGCUGUCAACAUGCUG ........-------..((((......(((((((.((((.--------------.......))))...((((((((((....))))..))))))))))))).......)))) ( -25.22) >DroSim_CAF1 53536 91 + 1 AACCGAAA-------UCCAGCUUCCACCAGCCAACAUGCA--------------GCAUUCAGCAUUCAGUGCCAGUGACAACUUACUUUGGCAUUUGGCUGUCAACAUGCUG ........-------..((((......(((((((.((((.--------------.......))))...((((((((((....))))..))))))))))))).......)))) ( -25.22) >DroYak_CAF1 58344 112 + 1 AACCAAAAUCCAACAUCCAACAUCCAACAUCCAACAUGCAGCAUUCAGCAUUCCGCAUUCAGCAUUCAGUACCAGUGACAACUUACUUUGGCAUUUGGCUGUCAGCAUGCUG ......................................((((((...((.....)).....((...((((...((((.(((......))).))))..))))...)))))))) ( -17.10) >consensus AACCAAAA_______UCCAGCUUCCACCAGCCAACAUGCA______________GCAUUCAGCAUUCAGUGCCAGUGACAACUUACUUUGGCAUUUGGCUGUCAACAUGCUG .................((((......(((((((....................((.....)).....((((((((((....))))..))))))))))))).......)))) (-18.84 = -19.59 + 0.75)

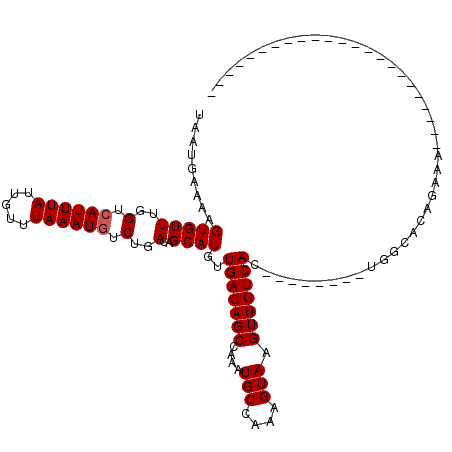
| Location | 14,535,820 – 14,535,918 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.22 |
| Mean single sequence MFE | -23.85 |
| Consensus MFE | -15.02 |
| Energy contribution | -15.18 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.560332 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14535820 98 - 22407834 UAAUGAAAAGUGUUUGGUCAUUUAUUGUUUAAAUGUCUGACAGCAUGUUGACAGCCAAAUGCCAAAGUAAGUUGUCAC--------UGGCACUGAAUGCUGAAUGC-------------- .........(((((..(.((((((.((((...((((......))))...))))((((..((.(((......))).)).--------))))..)))))))..)))))-------------- ( -21.30) >DroVir_CAF1 5691 91 - 1 UAAUGAAAAGUGUUUGGUCAUUUAUUGUUUAAAUGUCUGACAGCAUGUUGACAGCCAAAUGCCAAAGUAAGUUGUCAGAC------AGAAACAGAAA----------------------- .......(((((......))))).((((((...(((((((((((...(((.((......)).))).....))))))))))------).))))))...----------------------- ( -25.20) >DroEre_CAF1 62933 120 - 1 UAAUGAAAAGUGUUUGGUCAUUUAUUGUUUAAAUGUCUGACAGCAUGUUGACAGCCAAAUGCCAAAGUAAGUUGUCACUGCUACACUGGCACUGAAAGCUGAAUGCCGAAUGCUGCAUGC ..(((...(((((((((.((((((...((((..((((....((((.((.((((((....(((....))).)))))))))))).....)))).))))...))))))))))))))).))).. ( -32.50) >DroYak_CAF1 58384 112 - 1 UAAUGAAAAGUGUUUGGUCAUUUAUUGUUUAAAUGUCUGACAGCAUGCUGACAGCCAAAUGCCAAAGUAAGUUGUCAC--------UGGUACUGAAUGCUGAAUGCGGAAUGCUGAAUGC ........(((((((.(.((((((..(((((......(((((((.((((....((.....))...)))).))))))).--------......)))))..))))))).)))))))...... ( -26.12) >DroMoj_CAF1 7668 91 - 1 UAAUGAAAAGUGUUUGGUCAUUUAUUGUUUAAAUUUCUGACAGCAUGUUGACAGCCAAAUGCCAAAGUAAGUUGUCAGAC------AGAAACAGCAA----------------------- .......(((((......))))).((((((...(.(((((((((...(((.((......)).))).....))))))))).------).))))))...----------------------- ( -18.90) >DroAna_CAF1 5679 88 - 1 UAAUGAAAAGUGUUUGGUCAUUUAUUGUUUAAAUGUCUGACAGCAUGUUGACAGCCAAAUGCCAAAGUAAGUUGUCAG--------UGGCACAGCA------------------------ .........(((((....((((((.....)))))).((((((((...(((.((......)).))).....))))))))--------.)))))....------------------------ ( -19.10) >consensus UAAUGAAAAGUGUUUGGUCAUUUAUUGUUUAAAUGUCUGACAGCAUGUUGACAGCCAAAUGCCAAAGUAAGUUGUCAC________UGGCACAGAAA_______________________ .........(((((..(.((((((.....)))))).)..)..))))..(((((((....(((....))).)))))))........................................... (-15.02 = -15.18 + 0.17)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:36:18 2006