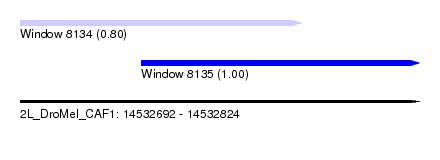
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,532,692 – 14,532,824 |
| Length | 132 |
| Max. P | 0.997461 |

| Location | 14,532,692 – 14,532,785 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 66.99 |
| Mean single sequence MFE | -26.21 |
| Consensus MFE | -11.03 |
| Energy contribution | -12.03 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.42 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.804222 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14532692 93 + 22407834 UAGCUUUUUGUUCAGGGGGGCCUUAUUCUCUGGCCCUUCUACU---------------------------UUCACUGGAGCCUUAUCCGAUUUCUUGGUGCCGAAACCAAGGAUGGAACC ..(((((.((...(((((((((.........)))))))))...---------------------------..))..)))))....((((..((((((((......))))))))))))... ( -32.70) >DroSec_CAF1 64684 93 + 1 UAAUUUUUUGUUCAGAGAGGGCUGCUUCUCUGGGACUUCUACU---------------------------UUCACUGGAGCCUUAUUCGAUUUCUUGGUGCCGAAACCAAGAAUAGAACC ............((((((((....))))))))((.((((....---------------------------......))))))...(((...((((((((......))))))))..))).. ( -27.80) >DroSim_CAF1 50042 93 + 1 UAGUUUUUUGUUCAGAGCGGGCUGCUUCUCUGGGACUUCUACU---------------------------UUCACUGGAGCCUUAUUCGAUUUCUUGGUGCCGAAACCAAGAAUAGAACC .........((((.(((..((..(((((..((..(.......)---------------------------..))..)))))))..)))...((((((((......))))))))..)))). ( -25.60) >DroEre_CAF1 59936 90 + 1 UAGUUUCUUGUUCAGACUGAAC---UUAUCUGAAACUUCUCCU---------------------------UUCACUGGAGCCUGAUCCGCUAUCUUGGUGCCGAAUCCAUGAAGAGAACC .((((((..((((.....))))---......))))))((((..---------------------------((((.((((.......(((......))).......))))))))))))... ( -19.44) >DroYak_CAF1 55129 81 + 1 UAGUUUUUUGUUCAGGG------------GUGAGCCCUCUCCU---------------------------UUAGCUGGAAUCUUAUCCGAUUUCUUGGUGCCGAAUCCAAGAAUAGAGCU .........((((..((------------(((((.....(((.---------------------------......)))..)))))))...(((((((........)))))))..)))). ( -21.60) >DroAna_CAF1 2104 120 + 1 CAGUUUCUUCUUCGAAGUUAUCGGUUUUGCUGAUUCUACUCCUUGCACGGGGCUAUUGUCCAUCUUGGACUUCACGGGAGCCUUCCCCAUUUUCUUUGGGCCCAAUCUGAAAAUGGACCC ......(((((..(((((((((((.....)))))...............((((....))))......))))))..)))))......((((((((.(((....)))...)))))))).... ( -30.10) >consensus UAGUUUUUUGUUCAGAGUGGGCUG_UUCUCUGAGACUUCUACU___________________________UUCACUGGAGCCUUAUCCGAUUUCUUGGUGCCGAAACCAAGAAUAGAACC ...........((((((..........))))))...(((((...................................(((......)))...((((((((......))))))))))))).. (-11.03 = -12.03 + 1.00)

| Location | 14,532,732 – 14,532,824 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 73.54 |
| Mean single sequence MFE | -24.66 |
| Consensus MFE | -9.51 |
| Energy contribution | -10.15 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.39 |
| SVM decision value | 2.86 |
| SVM RNA-class probability | 0.997461 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14532732 92 + 22407834 ACU---------------------------UUCACUGGAGCCUUAUCCGAUUUCUUGGUGCCGAAACCAAGGAUGGAACCCUCCACAACCAAAGCCAAAGCUAAAAUUAAUACCAAAGU (((---------------------------((...(((((.....((((..((((((((......))))))))))))...))))).......(((....)))............))))) ( -21.00) >DroSec_CAF1 64724 92 + 1 ACU---------------------------UUCACUGGAGCCUUAUUCGAUUUCUUGGUGCCGAAACCAAGAAUAGAACCCUCCACAACCAUAGCCAAAGCUAAAAUUAACAGCAAAGU (((---------------------------(((..(((((.....(((...((((((((......))))))))..)))..)))))......((((....)))).........).))))) ( -21.90) >DroSim_CAF1 50082 92 + 1 ACU---------------------------UUCACUGGAGCCUUAUUCGAUUUCUUGGUGCCGAAACCAAGAAUAGAACCCUCCACAACCAUAGCCAAAGCUAAAAUUAACAGCAAAGU (((---------------------------(((..(((((.....(((...((((((((......))))))))..)))..)))))......((((....)))).........).))))) ( -21.90) >DroYak_CAF1 55157 92 + 1 CCU---------------------------UUAGCUGGAAUCUUAUCCGAUUUCUUGGUGCCGAAUCCAAGAAUAGAGCUCUCCACAACCGCAGCCAAAGCUAAAAUUAACAGCAAAGU .((---------------------------((.((((((.(((........(((((((........))))))).)))..))...........(((....)))........)))))))). ( -17.50) >DroAna_CAF1 2144 119 + 1 CCUUGCACGGGGCUAUUGUCCAUCUUGGACUUCACGGGAGCCUUCCCCAUUUUCUUUGGGCCCAAUCUGAAAAUGGACCCCUCGACACCCGUGGCCAAUACAAGAACAAACAGCAAGGG ((((((...((((....)))).(((((....(((((((.((.....((((((((.(((....)))...)))))))).......).).)))))))......))))).......)))))). ( -41.00) >consensus ACU___________________________UUCACUGGAGCCUUAUCCGAUUUCUUGGUGCCGAAACCAAGAAUAGAACCCUCCACAACCAUAGCCAAAGCUAAAAUUAACAGCAAAGU ...................................(((((...........((((((((......)))))))).......))))).................................. ( -9.51 = -10.15 + 0.64)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:36:13 2006