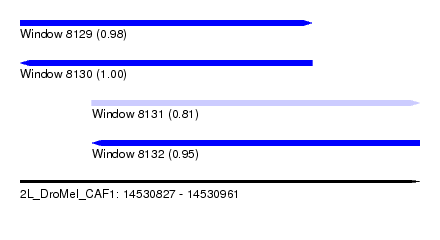
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,530,827 – 14,530,961 |
| Length | 134 |
| Max. P | 0.999897 |

| Location | 14,530,827 – 14,530,925 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 92.23 |
| Mean single sequence MFE | -31.78 |
| Consensus MFE | -30.86 |
| Energy contribution | -31.06 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.94 |
| SVM RNA-class probability | 0.983412 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14530827 98 + 22407834 UAAAUUAUGGUA--UAAAAAA-UGUAUUUUGCAUCGGCCGGGAAUCGAACCCAGGCCGCCCGCGUGGCAGGCGAGCAUUCUACCACUGAACCACCGAUGCU ........((((--((.....-))))))..(((((((..(((.......)))..((((((.((...)).)))).)).................))))))). ( -27.10) >DroSec_CAF1 62907 100 + 1 UAAAUUUUGGUAUUUAAAAAA-UGUAGUAUGCAUCGGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCUACCACUGAACCACCGAUGCU .....................-........((((((((((((.......)))))((((((.((...)).)))).)).................))))))). ( -32.20) >DroSim_CAF1 48261 100 + 1 UACAUUUUGGUAUUUAAAAAA-UAUAGUAUGCAUCGGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCUACCACUGAACCACCGAUGCU .....................-........((((((((((((.......)))))((((((.((...)).)))).)).................))))))). ( -32.20) >DroEre_CAF1 58063 101 + 1 AAAAGUAUGGAACUUAAAAAAAAGUCGUAUGCAUCGGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCUACCACUGAACCACCGAUGCU ....(((((..((((......)))))))))((((((((((((.......)))))((((((.((...)).)))).)).................))))))). ( -35.20) >DroYak_CAF1 53170 98 + 1 AAAAUUAUGGGAAUUAAAAAA---UCGUUUGCAUCGGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCUACCACUGAACCACCGAUGCU .....................---......((((((((((((.......)))))((((((.((...)).)))).)).................))))))). ( -32.20) >consensus UAAAUUAUGGUAUUUAAAAAA_UGUAGUAUGCAUCGGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCUACCACUGAACCACCGAUGCU ..............................((((((((((((.......)))))((((((.((...)).)))).)).................))))))). (-30.86 = -31.06 + 0.20)

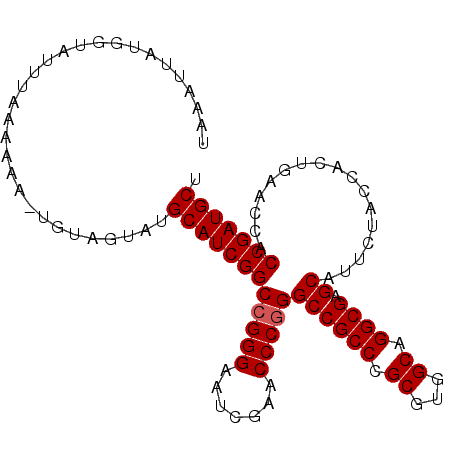
| Location | 14,530,827 – 14,530,925 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 92.23 |
| Mean single sequence MFE | -36.70 |
| Consensus MFE | -35.62 |
| Energy contribution | -35.46 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.97 |
| SVM decision value | 4.43 |
| SVM RNA-class probability | 0.999897 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14530827 98 - 22407834 AGCAUCGGUGGUUCAGUGGUAGAAUGCUCGCCUGCCACGCGGGCGGCCUGGGUUCGAUUCCCGGCCGAUGCAAAAUACA-UUUUUUA--UACCAUAAUUUA .(((((....((((.(((((((.........)))))))..))))((((.(((.......))))))))))))........-.......--............ ( -35.00) >DroSec_CAF1 62907 100 - 1 AGCAUCGGUGGUUCAGUGGUAGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUGCAUACUACA-UUUUUUAAAUACCAAAAUUUA ......((((........((((.((((((((((((...))))))((((.(((.......))))))))).)))).)))).-.........))))........ ( -37.77) >DroSim_CAF1 48261 100 - 1 AGCAUCGGUGGUUCAGUGGUAGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUGCAUACUAUA-UUUUUUAAAUACCAAAAUGUA .((((...((((......((((.((((((((((((...))))))((((.(((.......))))))))).)))).)))).-..........))))..)))). ( -37.13) >DroEre_CAF1 58063 101 - 1 AGCAUCGGUGGUUCAGUGGUAGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUGCAUACGACUUUUUUUUAAGUUCCAUACUUUU .(((((((..((((.......))))((.(((((((...)))))))))(((((.......))))))))))))....(((((......))))).......... ( -37.70) >DroYak_CAF1 53170 98 - 1 AGCAUCGGUGGUUCAGUGGUAGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUGCAAACGA---UUUUUUAAUUCCCAUAAUUUU .(((((((..((((.......))))((.(((((((...)))))))))(((((.......))))))))))))......---..................... ( -35.90) >consensus AGCAUCGGUGGUUCAGUGGUAGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUGCAUACUACA_UUUUUUAAAUACCAUAAUUUA .(((((((..((((.......))))((.(((((((...)))))))))(((((.......)))))))))))).............................. (-35.62 = -35.46 + -0.16)

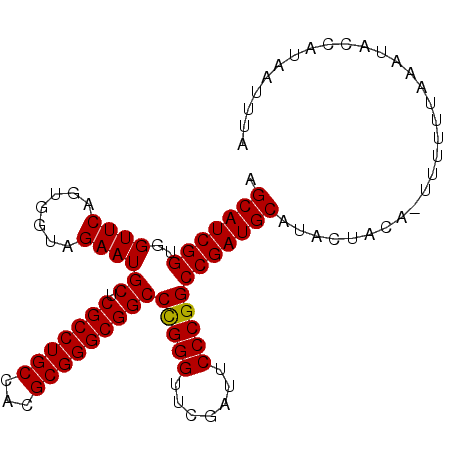
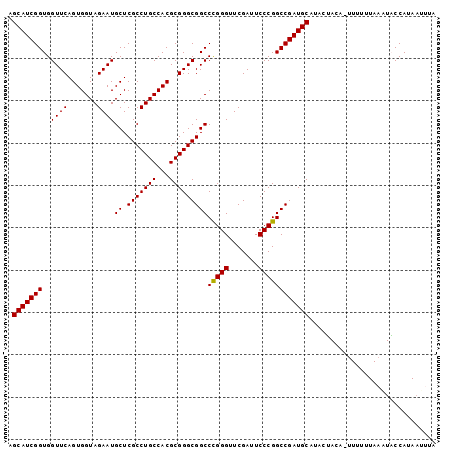
| Location | 14,530,851 – 14,530,961 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 89.13 |
| Mean single sequence MFE | -33.01 |
| Consensus MFE | -30.86 |
| Energy contribution | -31.06 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.813395 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14530851 110 + 22407834 UUUGCAUCGGCCGGGAAUCGAACCCAGGCCGCCCGCGUGGCAGGCGAGCAUUCUACCACUGAACCACCGAUGCUUCUUUAUUU-CUUGCGUCAGCAUUUUCCAAUAAUA---AU ...(((((((..(((.......)))..((((((.((...)).)))).)).................)))))))..........-..(((....))).............---.. ( -29.40) >DroSec_CAF1 62933 109 + 1 UAUGCAUCGGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCUACCACUGAACCACCGAUGCUUUU-UAGUU-UUUUCGUCAAAAUUUCCCAACAAUA---AU ...((((((((((((.......)))))((((((.((...)).)))).)).................)))))))....-.....-.........................---.. ( -32.20) >DroSim_CAF1 48287 109 + 1 UAUGCAUCGGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCUACCACUGAACCACCGAUGCUUCU-UAGUU-UUUGCGUCAAAAUUUCCCAACUAUA---AU ...((((((((((((.......)))))((((((.((...)).)))).)).................)))))))....-.....-.........................---.. ( -32.20) >DroEre_CAF1 58090 113 + 1 UAUGCAUCGGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCUACCACUGAACCACCGAUGCUUCA-UAUUUCGUUGCGUCAAAAUUUCCAAAUAAAAGCGAC ...((((((((((((.......)))))((((((.((...)).)))).)).................)))))))....-......(((((....................))))) ( -35.75) >DroYak_CAF1 53194 113 + 1 UUUGCAUCGGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCUACCACUGAACCACCGAUGCUUCU-UAUUUUCUUUGGUCAAAAUUACCGAACAUAAACUAC ...((((((((((((.......)))))((((((.((...)).)))).)).................)))))))....-.......((((((.......)))))).......... ( -35.50) >consensus UAUGCAUCGGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCUACCACUGAACCACCGAUGCUUCU_UAUUU_CUUGCGUCAAAAUUUCCCAACAAUA___AU ...((((((((((((.......)))))((((((.((...)).)))).)).................)))))))......................................... (-30.86 = -31.06 + 0.20)

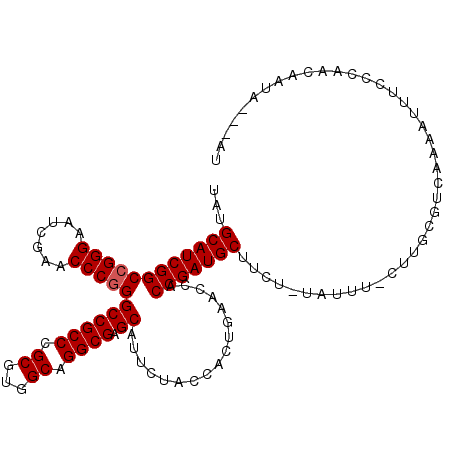
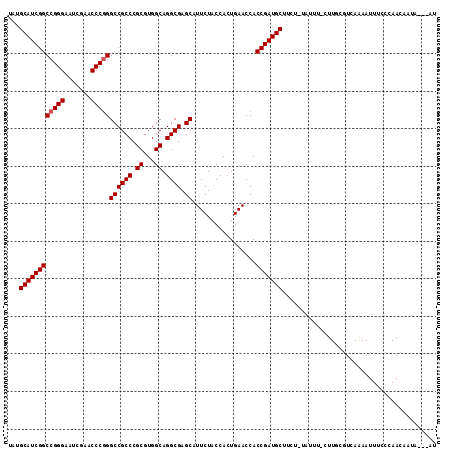
| Location | 14,530,851 – 14,530,961 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 89.13 |
| Mean single sequence MFE | -39.32 |
| Consensus MFE | -35.62 |
| Energy contribution | -35.46 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.952586 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14530851 110 - 22407834 AU---UAUUAUUGGAAAAUGCUGACGCAAG-AAAUAAAGAAGCAUCGGUGGUUCAGUGGUAGAAUGCUCGCCUGCCACGCGGGCGGCCUGGGUUCGAUUCCCGGCCGAUGCAAA .(---((((((((((...((((((.((...-..........)).)))))).)))))))))))..(((((((((((...))))))((((.(((.......))))))))).))).. ( -42.42) >DroSec_CAF1 62933 109 - 1 AU---UAUUGUUGGGAAAUUUUGACGAAAA-AACUA-AAAAGCAUCGGUGGUUCAGUGGUAGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUGCAUA ..---..((((..((....))..))))...-.....-....(((((((..((((.......))))((.(((((((...)))))))))(((((.......))))))))))))... ( -39.50) >DroSim_CAF1 48287 109 - 1 AU---UAUAGUUGGGAAAUUUUGACGCAAA-AACUA-AGAAGCAUCGGUGGUUCAGUGGUAGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUGCAUA ..---....((..((....))..)).....-.....-....(((((((..((((.......))))((.(((((((...)))))))))(((((.......))))))))))))... ( -38.10) >DroEre_CAF1 58090 113 - 1 GUCGCUUUUAUUUGGAAAUUUUGACGCAACGAAAUA-UGAAGCAUCGGUGGUUCAGUGGUAGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUGCAUA .................((((((......)))))).-....(((((((..((((.......))))((.(((((((...)))))))))(((((.......))))))))))))... ( -37.70) >DroYak_CAF1 53194 113 - 1 GUAGUUUAUGUUCGGUAAUUUUGACCAAAGAAAAUA-AGAAGCAUCGGUGGUUCAGUGGUAGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUGCAAA ....(((((.((((((.......)))...))).)))-))..(((((((..((((.......))))((.(((((((...)))))))))(((((.......))))))))))))... ( -38.90) >consensus AU___UAUUGUUGGGAAAUUUUGACGCAAA_AAAUA_AGAAGCAUCGGUGGUUCAGUGGUAGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUGCAUA .........................................(((((((..((((.......))))((.(((((((...)))))))))(((((.......))))))))))))... (-35.62 = -35.46 + -0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:36:11 2006