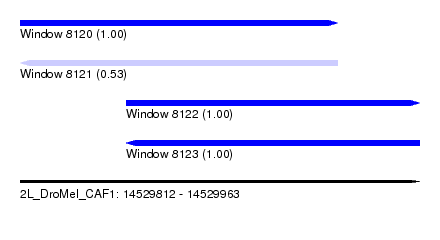
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,529,812 – 14,529,963 |
| Length | 151 |
| Max. P | 0.999971 |

| Location | 14,529,812 – 14,529,932 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.46 |
| Mean single sequence MFE | -36.75 |
| Consensus MFE | -32.35 |
| Energy contribution | -32.69 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.00 |
| Structure conservation index | 0.88 |
| SVM decision value | 3.02 |
| SVM RNA-class probability | 0.998165 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14529812 120 + 22407834 UUCGGUCGGCCUCAUGCCUCCUUAAGUUGACGUGUUACGCUGUCAGCAUGGAGAAAACCUCAAUUUCAAACUGUCAGUUUAUAUGUCAGCUCCAACUGACAGUUCUCCAUGUUGUCGCUG ..(((.((((..((((.((((....(((((((((...))).))))))..))))...............(((((((((((.....(....)...)))))))))))...))))..))))))) ( -36.70) >DroPse_CAF1 67266 108 + 1 UUC--UCGGC----------CUUAAGUUGACGUGUUACGCUGUCAGCAUGGAGAAAACCUCAAUUUCAAACUGUCAGUUUAUAUGUCAGCUUCAACUGACAGUUCUCCAUGUUGGCGCUG ...--(((((----------.....)))))........((.(((((((((((((...............((((((((((.....(....)...))))))))))))))))))))))))).. ( -35.76) >DroSim_CAF1 47209 120 + 1 UUCGGUCGGCCUCAUGCCUCCUUAAGUUGACGUGUUACGCUGUCAGCAUGGAGAAAACCUCAAUUUCAAACUGUCAGUUUAUAUGUCAGCUCCAACUGACAGUUCUCCAUGUUGUCGCUG ..(((.((((..((((.((((....(((((((((...))).))))))..))))...............(((((((((((.....(....)...)))))))))))...))))..))))))) ( -36.70) >DroEre_CAF1 57020 120 + 1 UUCGGUCGGCCUCAUGCCUCCUUAAGUUGACGUGUUACGCUGUCAGCAUGGAGAAAACCUCAAUUUCAAACUGUCAGUUUAUAUGUCAGCUCCAACUGACAGUUCUCCAUGUUGUCGCCG ..(((.((((..((((.((((....(((((((((...))).))))))..))))...............(((((((((((.....(....)...)))))))))))...))))..))))))) ( -38.90) >DroYak_CAF1 52127 120 + 1 UUCGGUCGGCCUCAUGCCUCCUUAAGUUGACGUGUUACGCUGUCAGCAUGGAGAAAACCUCAAUUUCAAACUGUCAGUUUAUAUGUCAGCUCCAACUGACAGUUCUCCAUGUUGUCGCUG ..(((.((((..((((.((((....(((((((((...))).))))))..))))...............(((((((((((.....(....)...)))))))))))...))))..))))))) ( -36.70) >DroPer_CAF1 71003 108 + 1 UUC--UCGGC----------CUUAAGUUGACGUGUUACGCUGUCAGCAUGGAGAAAACCUCAAUUUCAAACUGUCAGUUUAUAUGUCAGCUUCAACUGACAGUUCUCCAUGUUGGCGCUG ...--(((((----------.....)))))........((.(((((((((((((...............((((((((((.....(....)...))))))))))))))))))))))))).. ( -35.76) >consensus UUCGGUCGGCCUCAUGCCUCCUUAAGUUGACGUGUUACGCUGUCAGCAUGGAGAAAACCUCAAUUUCAAACUGUCAGUUUAUAUGUCAGCUCCAACUGACAGUUCUCCAUGUUGUCGCUG ....((((((...............)))))).......((.(.(((((((((((...............((((((((((.....(....)...))))))))))))))))))))).))).. (-32.35 = -32.69 + 0.33)

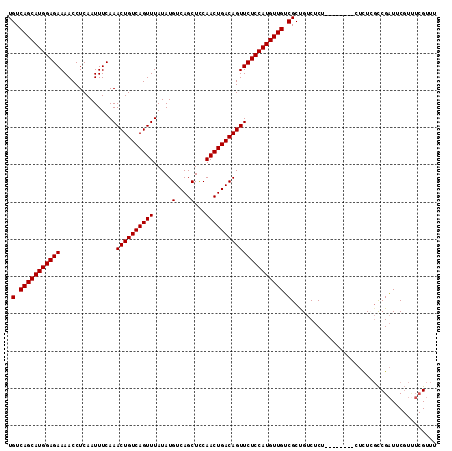
| Location | 14,529,812 – 14,529,932 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.46 |
| Mean single sequence MFE | -33.69 |
| Consensus MFE | -28.04 |
| Energy contribution | -28.04 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.83 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.530582 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14529812 120 - 22407834 CAGCGACAACAUGGAGAACUGUCAGUUGGAGCUGACAUAUAAACUGACAGUUUGAAAUUGAGGUUUUCUCCAUGCUGACAGCGUAACACGUCAACUUAAGGAGGCAUGAGGCCGACCGAA ..((..((.((((((((((((((((((..............))))))))))))((((.......)))))))))).))...)).................((.(((.....)))..))... ( -34.54) >DroPse_CAF1 67266 108 - 1 CAGCGCCAACAUGGAGAACUGUCAGUUGAAGCUGACAUAUAAACUGACAGUUUGAAAUUGAGGUUUUCUCCAUGCUGACAGCGUAACACGUCAACUUAAG----------GCCGA--GAA ..(.(((..((((((((((((((((((..............))))))))))))((((.......)))))))))).((((..........))))......)----------)))..--... ( -31.34) >DroSim_CAF1 47209 120 - 1 CAGCGACAACAUGGAGAACUGUCAGUUGGAGCUGACAUAUAAACUGACAGUUUGAAAUUGAGGUUUUCUCCAUGCUGACAGCGUAACACGUCAACUUAAGGAGGCAUGAGGCCGACCGAA ..((..((.((((((((((((((((((..............))))))))))))((((.......)))))))))).))...)).................((.(((.....)))..))... ( -34.54) >DroEre_CAF1 57020 120 - 1 CGGCGACAACAUGGAGAACUGUCAGUUGGAGCUGACAUAUAAACUGACAGUUUGAAAUUGAGGUUUUCUCCAUGCUGACAGCGUAACACGUCAACUUAAGGAGGCAUGAGGCCGACCGAA ((((..((.((((((((((((((((((..............))))))))))))((((.......)))))))))).)).........((.(((..(....)..))).))..))))...... ( -35.84) >DroYak_CAF1 52127 120 - 1 CAGCGACAACAUGGAGAACUGUCAGUUGGAGCUGACAUAUAAACUGACAGUUUGAAAUUGAGGUUUUCUCCAUGCUGACAGCGUAACACGUCAACUUAAGGAGGCAUGAGGCCGACCGAA ..((..((.((((((((((((((((((..............))))))))))))((((.......)))))))))).))...)).................((.(((.....)))..))... ( -34.54) >DroPer_CAF1 71003 108 - 1 CAGCGCCAACAUGGAGAACUGUCAGUUGAAGCUGACAUAUAAACUGACAGUUUGAAAUUGAGGUUUUCUCCAUGCUGACAGCGUAACACGUCAACUUAAG----------GCCGA--GAA ..(.(((..((((((((((((((((((..............))))))))))))((((.......)))))))))).((((..........))))......)----------)))..--... ( -31.34) >consensus CAGCGACAACAUGGAGAACUGUCAGUUGGAGCUGACAUAUAAACUGACAGUUUGAAAUUGAGGUUUUCUCCAUGCUGACAGCGUAACACGUCAACUUAAGGAGGCAUGAGGCCGACCGAA ..((..((.((((((((((((((((((..............))))))))))))((((.......)))))))))).))...(((.....)))...................))........ (-28.04 = -28.04 + -0.00)

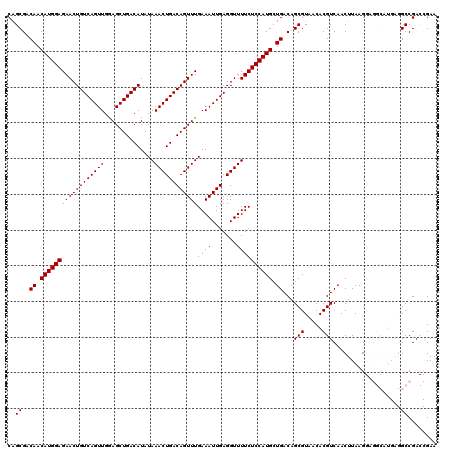
| Location | 14,529,852 – 14,529,963 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 90.23 |
| Mean single sequence MFE | -30.90 |
| Consensus MFE | -28.14 |
| Energy contribution | -28.14 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.84 |
| Structure conservation index | 0.91 |
| SVM decision value | 5.06 |
| SVM RNA-class probability | 0.999971 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14529852 111 + 22407834 UGUCAGCAUGGAGAAAACCUCAAUUUCAAACUGUCAGUUUAUAUGUCAGCUCCAACUGACAGUUCUCCAUGUUGUCGCUGUCUCUCU----CUCUCUCGCCGAUUCGUUUCGUUU .(.(((((((((((...............((((((((((.....(....)...))))))))))))))))))))).)...........----.........(((......)))... ( -29.76) >DroPse_CAF1 67294 102 + 1 UGUCAGCAUGGAGAAAACCUCAAUUUCAAACUGUCAGUUUAUAUGUCAGCUUCAACUGACAGUUCUCCAUGUUGGCGCUGUCUCUU--------UUUCUUCGUUUC-----GUUU .(((((((((((((...............((((((((((.....(....)...)))))))))))))))))))))))..........--------............-----.... ( -31.66) >DroSec_CAF1 61910 107 + 1 UGUCAGCAUGGAGAAAACCUCAAUUUCAAACUGUCAGUUUAUAUGUCAGCUCCAACUGACAGUUCUCCAUGUUGUCGCUGUCUCC--------CUCUAGCCGAUUCGUUUCGUUU .(.(((((((((((...............((((((((((.....(....)...))))))))))))))))))))).)((((.....--------...))))(((......)))... ( -32.06) >DroSim_CAF1 47249 107 + 1 UGUCAGCAUGGAGAAAACCUCAAUUUCAAACUGUCAGUUUAUAUGUCAGCUCCAACUGACAGUUCUCCAUGUUGUCGCUGUCUCC--------CUCUAGCCGAUUCGUUUCGUUU .(.(((((((((((...............((((((((((.....(....)...))))))))))))))))))))).)((((.....--------...))))(((......)))... ( -32.06) >DroEre_CAF1 57060 115 + 1 UGUCAGCAUGGAGAAAACCUCAAUUUCAAACUGUCAGUUUAUAUGUCAGCUCCAACUGACAGUUCUCCAUGUUGUCGCCGUCUCUCUCUUUCUCUCUCGCCGAUUCGUCUCGUUU .(.(((((((((((...............((((((((((.....(....)...))))))))))))))))))))).)........................(((......)))... ( -28.96) >consensus UGUCAGCAUGGAGAAAACCUCAAUUUCAAACUGUCAGUUUAUAUGUCAGCUCCAACUGACAGUUCUCCAUGUUGUCGCUGUCUCU________CUCUCGCCGAUUCGUUUCGUUU .(.(((((((((((...............((((((((((.....(....)...))))))))))))))))))))).)....................................... (-28.14 = -28.14 + 0.00)

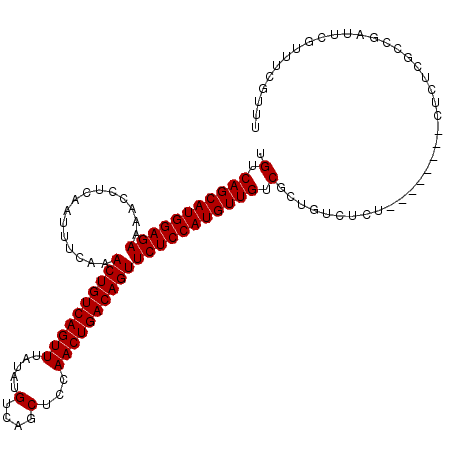
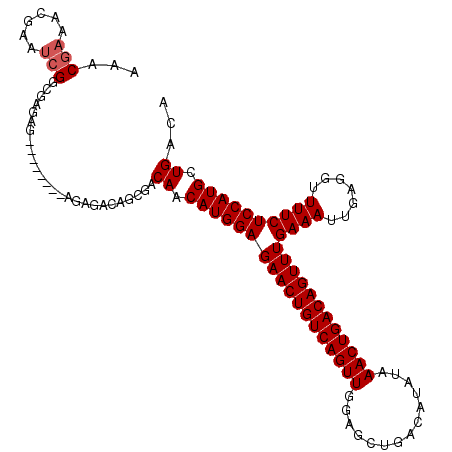
| Location | 14,529,852 – 14,529,963 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 90.23 |
| Mean single sequence MFE | -30.16 |
| Consensus MFE | -25.70 |
| Energy contribution | -26.10 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.85 |
| SVM decision value | 2.90 |
| SVM RNA-class probability | 0.997635 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14529852 111 - 22407834 AAACGAAACGAAUCGGCGAGAGAG----AGAGAGACAGCGACAACAUGGAGAACUGUCAGUUGGAGCUGACAUAUAAACUGACAGUUUGAAAUUGAGGUUUUCUCCAUGCUGACA ............((((((.(.(((----(((.........((..((....((((((((((((..............)))))))))))).....))..))))))))).)))))).. ( -29.24) >DroPse_CAF1 67294 102 - 1 AAAC-----GAAACGAAGAAA--------AAGAGACAGCGCCAACAUGGAGAACUGUCAGUUGAAGCUGACAUAUAAACUGACAGUUUGAAAUUGAGGUUUUCUCCAUGCUGACA ....-----............--------..........(.((.((((((((((((((((((..............))))))))))))((((.......)))))))))).)).). ( -25.54) >DroSec_CAF1 61910 107 - 1 AAACGAAACGAAUCGGCUAGAG--------GGAGACAGCGACAACAUGGAGAACUGUCAGUUGGAGCUGACAUAUAAACUGACAGUUUGAAAUUGAGGUUUUCUCCAUGCUGACA ............(((((.....--------(((((.(((..(((......((((((((((((..............))))))))))))....)))..))).)))))..))))).. ( -32.44) >DroSim_CAF1 47249 107 - 1 AAACGAAACGAAUCGGCUAGAG--------GGAGACAGCGACAACAUGGAGAACUGUCAGUUGGAGCUGACAUAUAAACUGACAGUUUGAAAUUGAGGUUUUCUCCAUGCUGACA ............(((((.....--------(((((.(((..(((......((((((((((((..............))))))))))))....)))..))).)))))..))))).. ( -32.44) >DroEre_CAF1 57060 115 - 1 AAACGAGACGAAUCGGCGAGAGAGAAAGAGAGAGACGGCGACAACAUGGAGAACUGUCAGUUGGAGCUGACAUAUAAACUGACAGUUUGAAAUUGAGGUUUUCUCCAUGCUGACA ............((((((.(.(((((((........(....)..((....((((((((((((..............)))))))))))).....))...)))))))).)))))).. ( -31.14) >consensus AAACGAAACGAAUCGGCGAGAG________AGAGACAGCGACAACAUGGAGAACUGUCAGUUGGAGCUGACAUAUAAACUGACAGUUUGAAAUUGAGGUUUUCUCCAUGCUGACA ...(((......)))..........................((.((((((((((((((((((..............))))))))))))((((.......)))))))))).))... (-25.70 = -26.10 + 0.40)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:36:03 2006