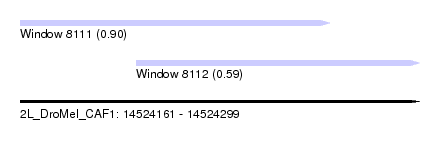
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,524,161 – 14,524,299 |
| Length | 138 |
| Max. P | 0.895475 |

| Location | 14,524,161 – 14,524,268 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 92.75 |
| Mean single sequence MFE | -25.25 |
| Consensus MFE | -22.95 |
| Energy contribution | -22.95 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.895475 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
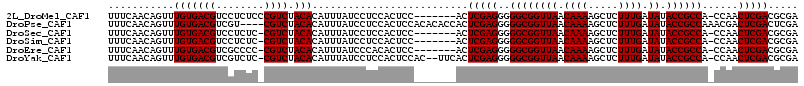
>2L_DroMel_CAF1 14524161 107 + 22407834 UUUCAACAGUUUGUGACGUCCUCUCCCGUCUACACAUUUAUCCUCCACUCC-------ACUCGAGGGGGCGGUUAACAAAAGCUCUUUGAUAUACCGCCA-CCAACUCGACGCGA ...........(((((((........)))).))).................-------..(((((((((((((((.((((.....)))).)).)))))).-))..)))))..... ( -25.30) >DroPse_CAF1 60587 111 + 1 UUUCAACAGUUUGUGACGUCGU----CGUCUACACAUUUAUCCUCCACUCCACACACCACUCGAGGGGGCGGUUAACAAAAGCUCUUUGAUAUACCGCCAAACGACUCGACUCGA .............(((.((((.----.(((..........(((((.................)))))((((((((.((((.....)))).)).))))))....))).))))))). ( -25.53) >DroSec_CAF1 56036 106 + 1 UUUCAACAGUUUGUGACGUCCUCUC-CGUCUACACAUUUAUCCUCCACUCC-------ACUCGAGGGGGCGGUUAACAAAAGCUCUUUGAUAUACCGCCA-CCAACUCGACGCGA ...........(((((((.......-)))).))).................-------..(((((((((((((((.((((.....)))).)).)))))).-))..)))))..... ( -25.00) >DroSim_CAF1 41414 106 + 1 UUUCAACAGUUUGUGACGUCCUCUC-CGUCUACACAUUUAUCCUCCACUCC-------ACUCGAGGGGGCGGUUAACAAAAGCUCUUUGAUAUACCGCCA-CCAACUCGACGCGA ...........(((((((.......-)))).))).................-------..(((((((((((((((.((((.....)))).)).)))))).-))..)))))..... ( -25.00) >DroEre_CAF1 52168 106 + 1 UUUCAACAGUUUGUGACGUCGCCCC-CGUCUACACAUUUAUCCCACACUCC-------ACUCGAGGGGGCGGUUAACAAAAGCUCUUUGAUAUACCGCCA-CCAACUCGACGCGA ...........(((((((.......-)))).))).................-------..(((((((((((((((.((((.....)))).)).)))))).-))..)))))..... ( -25.00) >DroYak_CAF1 46568 111 + 1 UUUCAACAGUUUGUGACGUCGUCUC-CGUCUACACAUUUAUCCUCCACUCCAC--UUCACUCGAGGGGGCGGUUAACAAAAGCUCUUUGAUAUACCGCCA-CCAACUCGACGCGA ...........(((((((.......-)))).)))...................--.((.((((((((((((((((.((((.....)))).)).)))))).-))..))))).).)) ( -25.70) >consensus UUUCAACAGUUUGUGACGUCCUCUC_CGUCUACACAUUUAUCCUCCACUCC_______ACUCGAGGGGGCGGUUAACAAAAGCUCUUUGAUAUACCGCCA_CCAACUCGACGCGA ...........(((((((........)))).)))..........................(((((..((((((((.((((.....)))).)).))))))......)))))..... (-22.95 = -22.95 + -0.00)

| Location | 14,524,201 – 14,524,299 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 90.79 |
| Mean single sequence MFE | -29.32 |
| Consensus MFE | -22.03 |
| Energy contribution | -22.20 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.10 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.590992 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14524201 98 + 22407834 UCCUCCACUCC-------ACUCGAGGGGGCGGUUAACAAAAGCUCUUUGAUAUACCGCCA-CCAACUCGACGCGACUC-------GACUUUCGAGACAUG--CUACCAACACUUC ...........-------..(((((((((((((((.((((.....)))).)).)))))).-))..))))).(((.(((-------(.....))))...))--)............ ( -26.40) >DroPse_CAF1 60623 115 + 1 UCCUCCACUCCACACACCACUCGAGGGGGCGGUUAACAAAAGCUCUUUGAUAUACCGCCAAACGACUCGACUCGACUCGAGUCGAGACUUUCAAGACAUGCGCUACCAACACUUG ...................((.(((((((((((((.((((.....)))).)).))))))......(((((((((...))))))))).))))).)).................... ( -32.10) >DroSec_CAF1 56075 98 + 1 UCCUCCACUCC-------ACUCGAGGGGGCGGUUAACAAAAGCUCUUUGAUAUACCGCCA-CCAACUCGACGCGACUC-------GACUUUCGAGACAUG--CUACCAACACUUC ...........-------..(((((((((((((((.((((.....)))).)).)))))).-))..))))).(((.(((-------(.....))))...))--)............ ( -26.40) >DroSim_CAF1 41453 98 + 1 UCCUCCACUCC-------ACUCGAGGGGGCGGUUAACAAAAGCUCUUUGAUAUACCGCCA-CCAACUCGACGCGACUC-------GACUUUCGAGACAUG--CUACCAACACUUC ...........-------..(((((((((((((((.((((.....)))).)).)))))).-))..))))).(((.(((-------(.....))))...))--)............ ( -26.40) >DroEre_CAF1 52207 98 + 1 UCCCACACUCC-------ACUCGAGGGGGCGGUUAACAAAAGCUCUUUGAUAUACCGCCA-CCAACUCGACGCGAGUC-------GACUUUCGAGACAUG--CUACCAACACUUC ...........-------.((((((((((((((((.((((.....)))).)).)))))).-.....(((((....)))-------)))))))))).....--............. ( -32.30) >DroYak_CAF1 46607 103 + 1 UCCUCCACUCCAC--UUCACUCGAGGGGGCGGUUAACAAAAGCUCUUUGAUAUACCGCCA-CCAACUCGACGCGAGUC-------GACUUUCGAGACAUG--CUACCAACACUUC .............--....((((((((((((((((.((((.....)))).)).)))))).-.....(((((....)))-------)))))))))).....--............. ( -32.30) >consensus UCCUCCACUCC_______ACUCGAGGGGGCGGUUAACAAAAGCUCUUUGAUAUACCGCCA_CCAACUCGACGCGACUC_______GACUUUCGAGACAUG__CUACCAACACUUC ...................((((((((((((((((.((((.....)))).)).)))))).......(((...)))............)))))))).................... (-22.03 = -22.20 + 0.17)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:35:52 2006