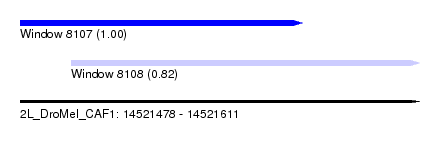
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,521,478 – 14,521,611 |
| Length | 133 |
| Max. P | 0.997283 |

| Location | 14,521,478 – 14,521,572 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 91.80 |
| Mean single sequence MFE | -23.04 |
| Consensus MFE | -20.88 |
| Energy contribution | -20.80 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.91 |
| SVM decision value | 2.83 |
| SVM RNA-class probability | 0.997283 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14521478 94 + 22407834 ---------CAUUGCAAGCAUUUGUU-UUGGCUAUAAAAUAUGAGCCAACACGGCACUUAAUUCUCAACGAAGCAUUCUGUGCCACAGCCCCAUGCACUGAAAA ---------((.((((.((....((.-((((((..........)))))).))(((((..(((.((......)).)))..)))))...))....)))).)).... ( -23.80) >DroSec_CAF1 53101 104 + 1 CAUUAACCCCAUUGCAAGCAUUUGUUUUUGGCUAUAAAAUAUGAGCCAACACGGCACUUAAUUCUCAACGAAGCAUUCGGUGCCACAGCCCCAUGCUCUGAAAU ..............(((((((((((..((((((..........))))))...((((((.(((.((......)).))).))))))))))....))))).)).... ( -22.10) >DroSim_CAF1 38747 103 + 1 CAUUAACCCCAUUGCAAGCAUUUGUU-UUGGCUAUAAAAUAUGAGCCAGCACGGCACUUAAUUCUCAACGAAGCAUUCGGUGCCACAGCCCCAUGCACUGAAAU .........((.((((.((....((.-((((((..........)))))).))((((((.(((.((......)).))).))))))...))....)))).)).... ( -25.10) >DroEre_CAF1 49667 104 + 1 CAUUAACCCCAUUGCGAGCAUUUGUUUUUGGCCAUAAAAUAUGAGCCAACUCGGCACUUAAUUCUCAACGAAGCAUUCUGUGCCACAGCCCCAUGCACUGAAAU .........((.((((.((........(((((((((...)))).)))))...(((((..(((.((......)).)))..)))))...))....)))).)).... ( -23.00) >DroYak_CAF1 43908 103 + 1 CAUUAACCCCAUUGCAAGCAUUUGUU-UUGGCUAUAAAAUAUGAGCCAACUCGGCACUUAAUUCCCAACGAAGCAUUCUGUGCCACAGUCCCAUGCACUGAAAC .........((.((((.(...((((.-((((((..........))))))...(((((....(((.....))).......)))))))))...).)))).)).... ( -21.20) >consensus CAUUAACCCCAUUGCAAGCAUUUGUU_UUGGCUAUAAAAUAUGAGCCAACACGGCACUUAAUUCUCAACGAAGCAUUCUGUGCCACAGCCCCAUGCACUGAAAU .........((.((((.(...((((..(((((((((...)))).)))))...(((((..(((.((......)).)))..)))))))))...).)))).)).... (-20.88 = -20.80 + -0.08)


| Location | 14,521,495 – 14,521,611 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 92.50 |
| Mean single sequence MFE | -25.65 |
| Consensus MFE | -21.39 |
| Energy contribution | -21.35 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.817340 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14521495 116 + 22407834 -UUGGCUAUAAAAUAUGAGCCAACACGGCACUUAAUUCUCAACGAAGCAUUCUGUGCCACAGCCCCAUGCACUGAAAAUAAGGCCGUAA-AAGUAUGUUGAAUGCACUCUAAAAAUAC -((((((..........)))))).(((((.((((.(((.....)))...(((.((((...........)))).)))..)))))))))..-..((((.....))))............. ( -24.90) >DroSec_CAF1 53127 117 + 1 UUUGGCUAUAAAAUAUGAGCCAACACGGCACUUAAUUCUCAACGAAGCAUUCGGUGCCACAGCCCCAUGCUCUGAAAUUAAGGCUGUAA-AAGUAUGUUGAAUGCACUCUAAAAAUAC .((((((..........)))))).(((((.(((((((.(((..((.((((..(((......)))..)))))))))))))))))))))..-..((((.....))))............. ( -29.30) >DroSim_CAF1 38773 116 + 1 -UUGGCUAUAAAAUAUGAGCCAGCACGGCACUUAAUUCUCAACGAAGCAUUCGGUGCCACAGCCCCAUGCACUGAAAUUAAGGCUGCAA-AAGUAUGUUGAAUGCACUCUAAAAAUAC -.(((((..........)))))(((.((((((.(((.((......)).))).)))))).(((((.................)))))...-............)))............. ( -25.53) >DroEre_CAF1 49693 118 + 1 UUUGGCCAUAAAAUAUGAGCCAACUCGGCACUUAAUUCUCAACGAAGCAUUCUGUGCCACAGCCCCAUGCACUGAAAUACAGGCUGUAAAAACCAUGUUGAAUGCAUUCUAAAAAUAC .(((((((((...)))).)))))...(((((..(((.((......)).)))..)))))((((((.................)))))).......((((.....))))........... ( -25.93) >DroYak_CAF1 43934 117 + 1 -UUGGCUAUAAAAUAUGAGCCAACUCGGCACUUAAUUCCCAACGAAGCAUUCUGUGCCACAGUCCCAUGCACUGAAACUAAGGCUGCAAGAAGUAUGUUGGAUGCAUUCUAAAAAUAC -...((............(((.....))).........((((((..((.(((((((((..(((..((.....))..)))..))).)).)))))).))))))..))............. ( -22.60) >consensus _UUGGCUAUAAAAUAUGAGCCAACACGGCACUUAAUUCUCAACGAAGCAUUCUGUGCCACAGCCCCAUGCACUGAAAUUAAGGCUGUAA_AAGUAUGUUGAAUGCACUCUAAAAAUAC .(((((((((...)))).)))))...(((((..(((.((......)).)))..))))).(((((.................)))))......((((.....))))............. (-21.39 = -21.35 + -0.04)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:35:48 2006