
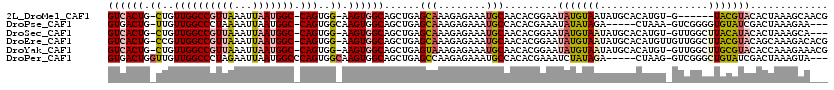
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,520,514 – 14,520,645 |
| Length | 131 |
| Max. P | 0.939175 |

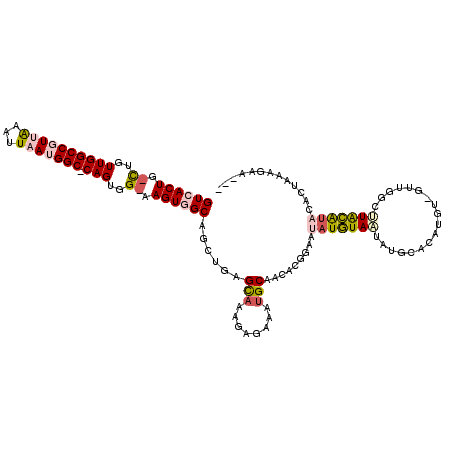
| Location | 14,520,514 – 14,520,624 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.57 |
| Mean single sequence MFE | -36.90 |
| Consensus MFE | -18.76 |
| Energy contribution | -19.49 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.25 |
| Mean z-score | -3.20 |
| Structure conservation index | 0.51 |
| SVM decision value | 1.28 |
| SVM RNA-class probability | 0.939175 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14520514 110 - 22407834 GUCACUG-CUGUUGGCCGUUAAAUUAAUGGC-CAGUGG-AAGUGGCAGCUGAGCAAAGAGAAAUGCAACACGGAAUAUGUAAUAUGCACAUGU-G------UACGUACACUAAAGCAACG ((((((.-(..((((((((((...)))))))-)))..)-.)))))).(((..(((........)))...(((..((((((.......))))))-.------..))).......))).... ( -37.50) >DroPse_CAF1 56643 109 - 1 GUGACUG-UUGUUGGCCCUAAAAUUAAUGGC-CAGUGGCAAGUGGCAGCUGAGCAAAGAGAAAUGCCACACGAAAUAUAUAGA-----CUAAA-GUCGGGGUGUAUCGACUAAAGAA--- .....((-(..((((((...........)))-)))..))).((((((.((........))...)))))).(((.((((...((-----(....-)))...)))).))).........--- ( -34.10) >DroSec_CAF1 52145 113 - 1 GUCACUG-CUGUUGGCCGUUAAAUUAAUGGC-CAGUGG-AAGUGGCAGCUGAGCAAAGAGAAAUGCAACACGGAAUAUGUAAUAUGCACAUGU-GUUGGCUUACAUACACUAAAGCA--- ((((((.-(..((((((((((...)))))))-)))..)-.)))))).(((..(((........))).....((..(((((((...(((....)-))....)))))))..))..))).--- ( -37.70) >DroEre_CAF1 48739 117 - 1 GUCACUG-CCGUUGGCCGUUAAAUUAAUGGC-CAGUGG-AAGUGGCAGCUGAGCAAAGAGAAAUGCAACACGGAAUAUGUAAUAUGCACAUGUUGUUGGCUUACGUACAGCAAAGACACG ((((((.-(((((((((((((...)))))))-))))))-.)))))).((((.((...(((....((((((.(..((((...))))...).))))))...)))..)).))))......... ( -40.80) >DroYak_CAF1 42907 116 - 1 GUCACUG-CUGUUGGCCGUUAAAUUAAUGGC-CAGUGG-AAGUGGCAGCUGAGUAAAGAGAAAUGCAACACGGAAUAUGUAAUAUGCACAUGU-GUUGGCUUGCGUACACCAAAGAAACG ((((((.-(..((((((((((...)))))))-)))..)-.))))))................((((((..(((.((((((.......))))))-.)))..)))))).............. ( -36.90) >DroPer_CAF1 60385 111 - 1 GUGACUGGUUGUUGGCCCUAGAAUUAAUGGCCCAGUGGCAAGUGGCAGCUGAGCCAAGAGAAAUGCCACACGAAAUCUAUAGA-----CUAAG-GUCGGGCUGUAUCGACUAAAGUA--- .....((((((.((((((((((.......(((....)))..((((((.((........))...))))))......))))..((-----(....-)))))))))...)))))).....--- ( -34.40) >consensus GUCACUG_CUGUUGGCCGUUAAAUUAAUGGC_CAGUGG_AAGUGGCAGCUGAGCAAAGAGAAAUGCAACACGGAAUAUGUAAUAUGCACAUGU_GUUGGCUUACAUACACUAAAGAA___ ((((((.((..((((((((((...))))))).)))..)).))))))......(((........))).........(((((((..................)))))))............. (-18.76 = -19.49 + 0.72)

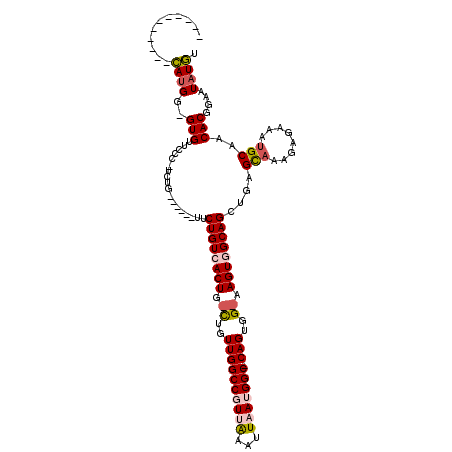
| Location | 14,520,547 – 14,520,645 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.61 |
| Mean single sequence MFE | -37.40 |
| Consensus MFE | -23.52 |
| Energy contribution | -24.25 |
| Covariance contribution | 0.73 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.82 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.889973 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14520547 98 - 22407834 -----------CAUGG-GUGUUCCC-UCUG------UUCUGUCACUG-CUGUUGGCCGUUAAAUUAAUGGC-CAGUGG-AAGUGGCAGCUGAGCAAAGAGAAAUGCAACACGGAAUAUGU -----------.....-(((((((.-..((------((((((((((.-(..((((((((((...)))))))-)))..)-.))))))))....(((........))))))).))))))).. ( -38.80) >DroPse_CAF1 56674 115 - 1 GGG-CAAUGCAUAUGG-GUGUUGUGUUCGGUUC-CUUACUGUGACUG-UUGUUGGCCCUAAAAUUAAUGGC-CAGUGGCAAGUGGCAGCUGAGCAAAGAGAAAUGCCACACGAAAUAUAU .((-((..((((....-))))..(((((((...-....((((.((((-(..((((((...........)))-)))..)).))).)))))))))))........))))............. ( -35.91) >DroSec_CAF1 52181 98 - 1 -----------CAUGG-GUGCUCCC-UCAG------UUCUGUCACUG-CUGUUGGCCGUUAAAUUAAUGGC-CAGUGG-AAGUGGCAGCUGAGCAAAGAGAAAUGCAACACGGAAUAUGU -----------...((-(....)))-((((------..((((((((.-(..((((((((((...)))))))-)))..)-.))))))))))))(((........))).............. ( -38.10) >DroEre_CAF1 48779 98 - 1 -----------CAUGG-GUGCUGCC-UCUG------UUCUGUCACUG-CCGUUGGCCGUUAAAUUAAUGGC-CAGUGG-AAGUGGCAGCUGAGCAAAGAGAAAUGCAACACGGAAUAUGU -----------((((.-(((.((((-((((------((((((((((.-(((((((((((((...)))))))-))))))-.)))))))...))))..))))....))).)))....)))). ( -38.40) >DroYak_CAF1 42946 98 - 1 -----------CAUGG-GUGUUCCC-UCUG------UUCUGUCACUG-CUGUUGGCCGUUAAAUUAAUGGC-CAGUGG-AAGUGGCAGCUGAGUAAAGAGAAAUGCAACACGGAAUAUGU -----------.....-(((((((.-..((------((((((((((.-(..((((((((((...)))))))-)))..)-.))))))))....(((........))))))).))))))).. ( -36.80) >DroPer_CAF1 60416 120 - 1 AGGCCAAUGCAUAUGGGGUGUUGUGUUCGGUUCCCUUACUGUGACUGGUUGUUGGCCCUAGAAUUAAUGGCCCAGUGGCAAGUGGCAGCUGAGCCAAGAGAAAUGCCACACGAAAUCUAU .(((((.......((((((.......(((((..(......)..)))))......)))))).......)))))..((((((..((((......)))).......))))))........... ( -36.36) >consensus ___________CAUGG_GUGUUCCC_UCUG______UUCUGUCACUG_CUGUUGGCCGUUAAAUUAAUGGC_CAGUGG_AAGUGGCAGCUGAGCAAAGAGAAAUGCAACACGGAAUAUGU ...........((((..(((..................((((((((.((..((((((((((...))))))).)))..)).))))))))....(((........)))..)))....)))). (-23.52 = -24.25 + 0.73)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:35:44 2006