
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,517,406 – 14,517,562 |
| Length | 156 |
| Max. P | 0.969157 |

| Location | 14,517,406 – 14,517,502 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 92.74 |
| Mean single sequence MFE | -35.32 |
| Consensus MFE | -28.00 |
| Energy contribution | -28.24 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.64 |
| SVM RNA-class probability | 0.969157 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14517406 96 - 22407834 AUGUCAUGCGACGCUGGAAAAGAUGACACCACUCUGCGGGAAACGGCCAGAG--------GAGCAGAGGGAUGAGCGUGAAAUACGAGUCCUUCGGCAUGUAGC ....(((((...(((((...........)).((((((.(....).).)))))--------.))).(((((((...((((...)))).))))))).))))).... ( -33.10) >DroSec_CAF1 48998 96 - 1 AUGUCAUGCGACGCUGGAAAAGAUGACACCACUCUGCGGGAAACGGCCAGAG--------GAGCCGAGGGAUGAGCGUGAAAUACGAGUCCUUCGGCAUGUAGC ..(((....)))((((.(.............((((((.(....).).)))))--------..((((((((((...((((...)))).)))))))))).).)))) ( -38.10) >DroSim_CAF1 34131 96 - 1 AUGUCAUGCGACGCUGGAAAAGAUGACACCACUCUGCGGGAAACGGCCAGAG--------GAGCCGAGGGAUGAGCGUGAAAUACGAGUCCUUCGGCAUGUAGC ..(((....)))((((.(.............((((((.(....).).)))))--------..((((((((((...((((...)))).)))))))))).).)))) ( -38.10) >DroEre_CAF1 42410 104 - 1 AUGUCAUCCGACGGUGGAAAAGAUGACACCACUCUGCGGGAAACAACCAGAGGAGCACAGGAGCAGAGGGAUGAGCGUGAAAUACGAGUCCUUCGGCAUGUAGC (((((.(((...((((..........)))).(((((..(....)...))))).......)))...(((((((...((((...)))).))))))))))))..... ( -35.30) >DroYak_CAF1 38965 96 - 1 AUGUCACCCGACGCUGGAAAAGAUGACACCACUCUGCGGGGAACGGCAGGAG--------GAGCAGAGGGAUGAGCGUGAAAUACGAGUCCUUCGGCAUGUAGC ..(((....)))((((.(..........((.(.((((.(....).))))).)--------).((.(((((((...((((...)))).))))))).)).).)))) ( -32.00) >consensus AUGUCAUGCGACGCUGGAAAAGAUGACACCACUCUGCGGGAAACGGCCAGAG________GAGCAGAGGGAUGAGCGUGAAAUACGAGUCCUUCGGCAUGUAGC ..(((....)))((((.(.............((((((.(....).)).))))..........((.(((((((...((((...)))).))))))).)).).)))) (-28.00 = -28.24 + 0.24)



| Location | 14,517,462 – 14,517,562 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.84 |
| Mean single sequence MFE | -33.23 |
| Consensus MFE | -22.25 |
| Energy contribution | -21.97 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.646210 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14517462 100 - 22407834 GGAGGUG-UCACAG--------CAAAACAGCUAC-----------CAGCGACAAUGACAACGGUUCCACAUUGUUGUCACAUGUCAUGCGACGCUGGAAAAGAUGACACCACUCUGCGGG ...((((-((((((--------(......))).(-----------((((.....((((((((((.....))))))))))...(((....))))))))....).))))))).......... ( -36.10) >DroPse_CAF1 52004 107 - 1 AAAUGUG-UCACAG--------CAAAACAGCUACCAAGCGACAGCCAGCGACAAUGACAACGGUUCCACAUUGUUGUCACAUGUCAUCCGACGCAGAAAAAGAUGACACGGCCAUG---- ...((((-((((((--------(......))).....(((...(....)((((.((((((((((.....))))))))))..))))......))).......).)))))))......---- ( -29.00) >DroSim_CAF1 34187 100 - 1 GGAGGUG-UCACAG--------CAAAACAGCUAC-----------CAGCGACAAUGACAACGGUUCCACAUUGUUGUCACAUGUCAUGCGACGCUGGAAAAGAUGACACCACUCUGCGGG ...((((-((((((--------(......))).(-----------((((.....((((((((((.....))))))))))...(((....))))))))....).))))))).......... ( -36.10) >DroEre_CAF1 42474 109 - 1 GGCGGGGAUCACAGCAAAGCAGCAAAACAGCUAC-----------CAGCGACAAUGACAACGGUUCCACAUUGUUGUCACAUGUCAUCCGACGGUGGAAAAGAUGACACCACUCUGCGGG .((((((.((...((...(.(((......))).)-----------..))((((.((((((((((.....))))))))))..))))....)).((((..........)))).))))))... ( -33.10) >DroYak_CAF1 39021 100 - 1 GGAGGUG-UCACAG--------CAAAACAGCUAC-----------CAGCGACAAUGACAACGGUUCCACAUUGUUGUCACAUGUCACCCGACGCUGGAAAAGAUGACACCACUCUGCGGG ...((((-((((((--------(......))).(-----------((((.....((((((((((.....))))))))))...(((....))))))))....).))))))).......... ( -36.10) >DroPer_CAF1 56987 107 - 1 AAAUGUG-UCACAG--------CAAAACAGCUACCAAGCGACAGCCAGCGACAAUGACAACGGUUCCACAUUGUUGUCACAUGUCAUCCGACGCAGAAAAAGAUGACACGGCCAUG---- ...((((-((((((--------(......))).....(((...(....)((((.((((((((((.....))))))))))..))))......))).......).)))))))......---- ( -29.00) >consensus GGAGGUG_UCACAG________CAAAACAGCUAC___________CAGCGACAAUGACAACGGUUCCACAUUGUUGUCACAUGUCAUCCGACGCUGGAAAAGAUGACACCACUCUGCGGG ...((((.((((.................(((..............)))((((.((((((((((.....))))))))))..))))................).))))))).......... (-22.25 = -21.97 + -0.28)

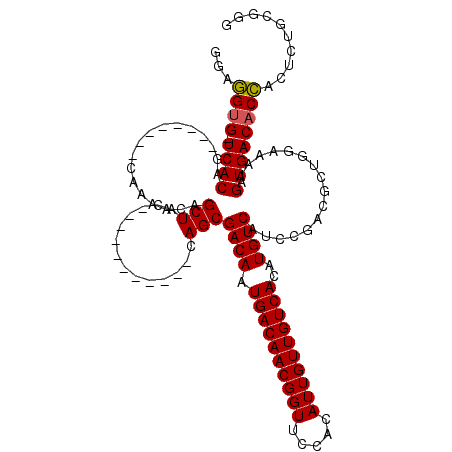
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:35:38 2006