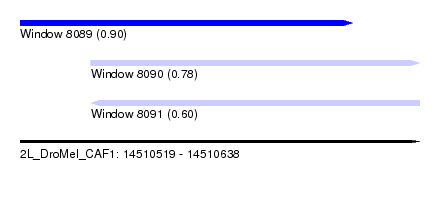
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,510,519 – 14,510,638 |
| Length | 119 |
| Max. P | 0.900458 |

| Location | 14,510,519 – 14,510,618 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 69.82 |
| Mean single sequence MFE | -34.77 |
| Consensus MFE | -19.73 |
| Energy contribution | -21.87 |
| Covariance contribution | 2.14 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.57 |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.900458 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
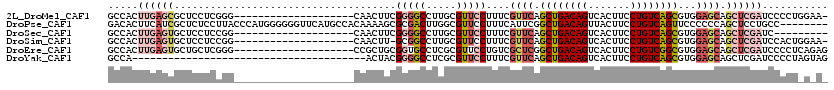
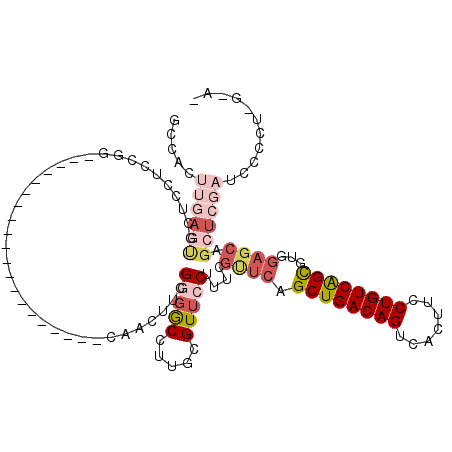
>2L_DroMel_CAF1 14510519 99 + 22407834 GCCACUUGAGCGCUCCUCGGG--------------------CAACUUCGGGGCCUUGCGUUCCUUUCGUUCAGCUGACAGUCACUUCCUGUCAGCGUGGAGCAGCUCGAUCCCCUGGAA- .(((.(((((((((((.((((--------------------(..(....).)))).................((((((((.......))))))))).))))).)))))).....)))..- ( -39.00) >DroPse_CAF1 40958 112 + 1 GACACUUCAUCGCUCUCCUUACCCAUGGGGGGUUCAUGCCACAAAAGCGCGACUUGGCGUUCCUUUCAUUCGGCUGACAGUUACUUCCUGUCAGUUCCCCCCAGCUCCUGCC-------- .........................(((((((.............(((((......)))))..........(((((((((.......))))))))))))))))((....)).-------- ( -32.10) >DroSec_CAF1 42101 91 + 1 GCCACUUGAGUGCUCCUCCGG--------------------CAACUUCGGGGCCUUGCGUUCCUUUCGUUCAGCUGACAGUCACUUCCUGUCAGCGUGGAGCAGCUCGAUC--------- .....(((((((((((.(.((--------------------(..(....).)))..................((((((((.......))))))))).)))))).)))))..--------- ( -35.00) >DroSim_CAF1 27234 98 + 1 GCCACUUGAGUGCUCCUCCGG--------------------CAACUU-GCGGCCUUGCGUUCCUUUCGUUCAGCUGACAGUCACUUCCUGUCAGCGUGGAGCAGCUCGAUCCACUGGAA- .(((.(((((((((((.(.((--------------------(.....-...)))..................((((((((.......))))))))).)))))).))))).....)))..- ( -34.40) >DroEre_CAF1 35500 100 + 1 GCCACUUGAGUGCUGCUCGGG--------------------CCGCUGCGGUGCCUCGCGUUCCUGUCGCUCGGCUGACAGUCACUUCCUGUCGGCGUGGAGCAGCUCGAUCCCCUCAGAG .....(((((.((((((((((--------------------(.(((((((....))))).....)).)))).((((((((.......))))))))...)))))))........))))).. ( -37.30) >DroYak_CAF1 31726 81 + 1 GCCA---------------------------------------ACUACGGGGCCUCGCGUUCCUUUCGUUCAGCUGACAGUCACUUCCUGUCAGCGUGGAGCAGCUCGAUCCCCUAGUAG ....---------------------------------------((((.((((..(((.(((......((((.((((((((.......))))))))...))))))).))).)))))))).. ( -30.80) >consensus GCCACUUGAGUGCUCCUCCGG____________________CAACUUCGGGGCCUUGCGUUCCUUUCGUUCAGCUGACAGUCACUUCCUGUCAGCGUGGAGCAGCUCGAUCCCCU_G_A_ .....((((((.....................................(((((.....)))))....((((.((((((((.......))))))))...)))).))))))........... (-19.73 = -21.87 + 2.14)
| Location | 14,510,540 – 14,510,638 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 68.89 |
| Mean single sequence MFE | -30.00 |
| Consensus MFE | -18.44 |
| Energy contribution | -19.08 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.775031 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
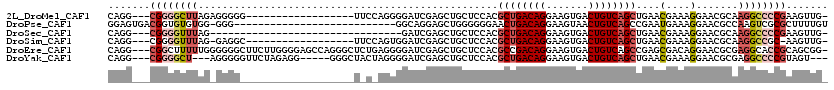
>2L_DroMel_CAF1 14510540 98 + 22407834 -CAACUUCGGGGCCUUGCGUUCCUUUCGUUCAGCUGACAGUCACUUCCUGUCAGCGUGGAGCAGCUCGAUCCCCUGGAA------------------CCCCCUCUAAGCCCCG---CCUG -......((((((...(((((((.........((((((((.......))))))))..))))).)).........((((.------------------.....)))).))))))---.... ( -33.30) >DroPse_CAF1 40998 92 + 1 ACAAAAGCGCGACUUGGCGUUCCUUUCAUUCGGCUGACAGUUACUUCCUGUCAGUUCCCCCCAGCUCCUGCC---------------------------CCC-CCACACACCGUCACUCC .....(((((......))))).........((((((((((.......))))))).........((....)).---------------------------...-.......)))....... ( -17.40) >DroSec_CAF1 42122 84 + 1 -CAACUUCGGGGCCUUGCGUUCCUUUCGUUCAGCUGACAGUCACUUCCUGUCAGCGUGGAGCAGCUCGAUC--------------------------------CUAAACCCCG---CCUG -......(((((..(((.(.((.....((((.((((((((.......))))))))...)))).....)).)--------------------------------.))).)))))---.... ( -26.50) >DroSim_CAF1 27255 96 + 1 -CAACUU-GCGGCCUUGCGUUCCUUUCGUUCAGCUGACAGUCACUUCCUGUCAGCGUGGAGCAGCUCGAUCCACUGGAA------------------GCCUC-CUAAACCCCG---CCUG -......-((((..(((.(...(((((.....((((((((.......))))))))(((((.(.....).))))).))))------------------)...)-.)))...)))---)... ( -29.00) >DroEre_CAF1 35521 116 + 1 -CCGCUGCGGUGCCUCGCGUUCCUGUCGCUCGGCUGACAGUCACUUCCUGUCGGCGUGGAGCAGCUCGAUCCCCUCAGAGCCCUGGCUCCCCAAGAAGCCCCCCAAAAAGCCG---CCUG -.....(((((((.(((((.......)))...((((((((.......))))))))(.(((((.((((..........))))....))))).)..)).))..........))))---)... ( -38.10) >DroYak_CAF1 31730 106 + 1 ---ACUACGGGGCCUCGCGUUCCUUUCGUUCAGCUGACAGUCACUUCCUGUCAGCGUGGAGCAGCUCGAUCCCCUAGUAGCCC-----CCUCUAGAACCCCCU---AGCCCCG---CCUG ---....((((((...(((((((.........((((((((.......))))))))..))))).))........((((.((...-----.))))))........---.))))))---.... ( -35.70) >consensus _CAACUUCGGGGCCUUGCGUUCCUUUCGUUCAGCUGACAGUCACUUCCUGUCAGCGUGGAGCAGCUCGAUCCCCU_G_A___________________CCCC_CUAAACCCCG___CCUG ........(((((.....)))))....((((.((((((((.......))))))))...)))).......................................................... (-18.44 = -19.08 + 0.64)

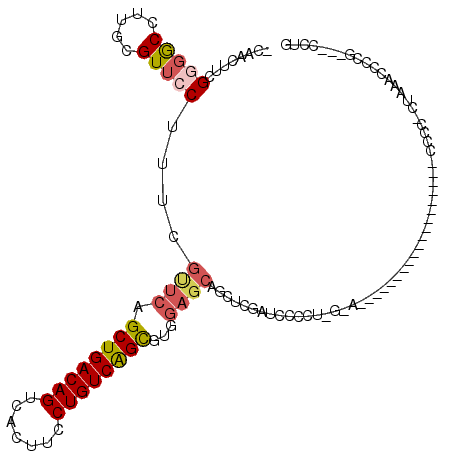
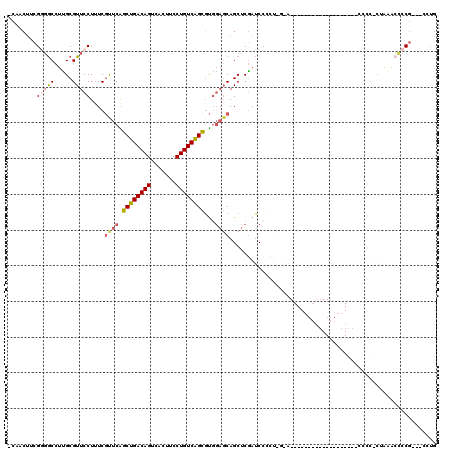
| Location | 14,510,540 – 14,510,638 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 68.89 |
| Mean single sequence MFE | -37.75 |
| Consensus MFE | -19.19 |
| Energy contribution | -20.38 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.604018 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14510540 98 - 22407834 CAGG---CGGGGCUUAGAGGGGG------------------UUCCAGGGGAUCGAGCUGCUCCACGCUGACAGGAAGUGACUGUCAGCUGAACGAAAGGAACGCAAGGCCCCGAAGUUG- ....---((((((((.....(.(------------------((((.((((.........))))..((((((((.......)))))))).........))))).).))))))))......- ( -42.00) >DroPse_CAF1 40998 92 - 1 GGAGUGACGGUGUGUGG-GGG---------------------------GGCAGGAGCUGGGGGGAACUGACAGGAAGUAACUGUCAGCCGAAUGAAAGGAACGCCAAGUCGCGCUUUUGU ...((((((((((....-...---------------------------(((....)))....((..(((((((.......)))))))))...........)))))..)))))........ ( -26.60) >DroSec_CAF1 42122 84 - 1 CAGG---CGGGGUUUAG--------------------------------GAUCGAGCUGCUCCACGCUGACAGGAAGUGACUGUCAGCUGAACGAAAGGAACGCAAGGCCCCGAAGUUG- ....---((((((((.(--------------------------------((.(.....).)))..((((((((.......))))))))....(....).......))))))))......- ( -30.50) >DroSim_CAF1 27255 96 - 1 CAGG---CGGGGUUUAG-GAGGC------------------UUCCAGUGGAUCGAGCUGCUCCACGCUGACAGGAAGUGACUGUCAGCUGAACGAAAGGAACGCAAGGCCGC-AAGUUG- ...(---(((.((((((-(((((------------------((((...)))...)))..))))..((((((((.......)))))))))))))........(....).))))-......- ( -34.60) >DroEre_CAF1 35521 116 - 1 CAGG---CGGCUUUUUGGGGGGCUUCUUGGGGAGCCAGGGCUCUGAGGGGAUCGAGCUGCUCCACGCCGACAGGAAGUGACUGUCAGCCGAGCGACAGGAACGCGAGGCACCGCAGCGG- ...(---(((.....(((((((((((....))))))..(((((.((.....))))))).)))))....(((((.......))))).(((..(((.......)))..))).)))).....- ( -47.00) >DroYak_CAF1 31730 106 - 1 CAGG---CGGGGCU---AGGGGGUUCUAGAGG-----GGGCUACUAGGGGAUCGAGCUGCUCCACGCUGACAGGAAGUGACUGUCAGCUGAACGAAAGGAACGCGAGGCCCCGUAGU--- ...(---(((((((---...(.(((((....(-----((((..((.(.....).))..)))))..((((((((.......)))))))).........))))).)..))))))))...--- ( -45.80) >consensus CAGG___CGGGGUUUAG_GGGG___________________U_C_AGGGGAUCGAGCUGCUCCACGCUGACAGGAAGUGACUGUCAGCUGAACGAAAGGAACGCAAGGCCCCGAAGUUG_ .......((((((((..................................................((((((((.......))))))))....(....).......))))))))....... (-19.19 = -20.38 + 1.20)
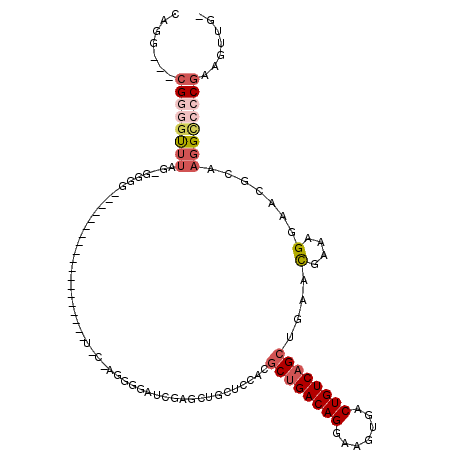
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:35:32 2006