| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,508,718 – 14,508,818 |
| Length | 100 |
| Max. P | 0.997374 |

| Location | 14,508,718 – 14,508,818 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.89 |
| Mean single sequence MFE | -33.50 |
| Consensus MFE | -30.82 |
| Energy contribution | -31.46 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.85 |
| SVM RNA-class probability | 0.997374 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14508718 100 + 22407834 CUGGGGAUCGGAUCGGUCCUGGAAUUGUAAAAUUGGGGUGUAACCCACUGUCGGCUUACACACUCAAGUCAAGCGACCAACGCCAUCCCCAU-C-------------------AACCGAA .(((((((.((...((((((.((.........(((((((((((.((......)).)))))).))))).)).)).))))....))))))))).-.-------------------....... ( -34.90) >DroSec_CAF1 40350 100 + 1 CUGGGGAUCGGAUCGGUCCUGGAAUUGUAAAAUUGGGGUGUAACCCACUGUCGGCUUACACACUCAAGUCAAGCGACCAACGCCAUCCCCAU-C-------------------AACCGAA .(((((((.((...((((((.((.........(((((((((((.((......)).)))))).))))).)).)).))))....))))))))).-.-------------------....... ( -34.90) >DroSim_CAF1 25053 100 + 1 CUGGGGAUCGGAUCGGUCCUGGAAUUGUAAAAUUGGGGUGUAACCCACUGUCGGCUUACACACUCAAGUCAAGCGACCAACGCCAUCCCCAU-G-------------------AACCGAA .(((((((.((...((((((.((.........(((((((((((.((......)).)))))).))))).)).)).))))....))))))))).-.-------------------....... ( -34.90) >DroEre_CAF1 33831 99 + 1 CUGG-CAUCGGAUCGGUCCUGGAAUUGUAAAAUUGGGGUGUAACCCACUGUCGGCUUACACACUCAAGUCAAGCGACCAACGCCAUCCCCAA-C-------------------AGCUGAA (((.-....((((.(((..(((..((((....(((((((((((.((......)).)))))).))))).....)))))))..)))))))....-)-------------------))..... ( -28.30) >DroYak_CAF1 29778 120 + 1 CUGGGGAUCGCAUCGGUCCUGGAAUUGUAAAAUUGGGGUGUAACCCACUGUCGGCUUACACACUCGAGUCAAGCGACCAACGCCAUCCCCCAUCAGCCUCACCAACAUAACCUAACUGAA ..((((((.((...((((((.((.........(((((((((((.((......)).)))))).))))).)).)).))))...)).))))))..((((...................)))). ( -34.51) >consensus CUGGGGAUCGGAUCGGUCCUGGAAUUGUAAAAUUGGGGUGUAACCCACUGUCGGCUUACACACUCAAGUCAAGCGACCAACGCCAUCCCCAU_C___________________AACCGAA .(((((((.((...((((((.((.........(((((((((((.((......)).)))))).))))).)).)).))))....)))))))))............................. (-30.82 = -31.46 + 0.64)

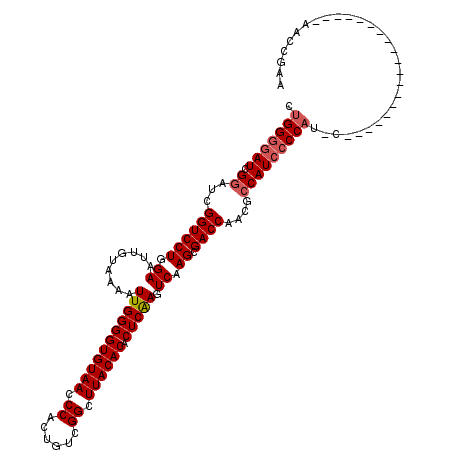
| Location | 14,508,718 – 14,508,818 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.89 |
| Mean single sequence MFE | -36.92 |
| Consensus MFE | -33.54 |
| Energy contribution | -34.14 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.894974 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14508718 100 - 22407834 UUCGGUU-------------------G-AUGGGGAUGGCGUUGGUCGCUUGACUUGAGUGUGUAAGCCGACAGUGGGUUACACCCCAAUUUUACAAUUCCAGGACCGAUCCGAUCCCCAG .......-------------------.-.(((((((.(.(((((((((((.....))))((((((.(((....))).))))))...................))))))).).))))))). ( -36.50) >DroSec_CAF1 40350 100 - 1 UUCGGUU-------------------G-AUGGGGAUGGCGUUGGUCGCUUGACUUGAGUGUGUAAGCCGACAGUGGGUUACACCCCAAUUUUACAAUUCCAGGACCGAUCCGAUCCCCAG .......-------------------.-.(((((((.(.(((((((((((.....))))((((((.(((....))).))))))...................))))))).).))))))). ( -36.50) >DroSim_CAF1 25053 100 - 1 UUCGGUU-------------------C-AUGGGGAUGGCGUUGGUCGCUUGACUUGAGUGUGUAAGCCGACAGUGGGUUACACCCCAAUUUUACAAUUCCAGGACCGAUCCGAUCCCCAG .......-------------------.-.(((((((.(.(((((((((((.....))))((((((.(((....))).))))))...................))))))).).))))))). ( -36.50) >DroEre_CAF1 33831 99 - 1 UUCAGCU-------------------G-UUGGGGAUGGCGUUGGUCGCUUGACUUGAGUGUGUAAGCCGACAGUGGGUUACACCCCAAUUUUACAAUUCCAGGACCGAUCCGAUG-CCAG .......-------------------.-.......((((((((((((((((((....))((((((.(((....))).))))))................))))..))).))))))-))). ( -29.00) >DroYak_CAF1 29778 120 - 1 UUCAGUUAGGUUAUGUUGGUGAGGCUGAUGGGGGAUGGCGUUGGUCGCUUGACUCGAGUGUGUAAGCCGACAGUGGGUUACACCCCAAUUUUACAAUUCCAGGACCGAUGCGAUCCCCAG .((((((...((((....))))))))))..((((((.((((((((((((((...)))))((((((.(((....))).))))))...................))))))))).)))))).. ( -46.10) >consensus UUCGGUU___________________G_AUGGGGAUGGCGUUGGUCGCUUGACUUGAGUGUGUAAGCCGACAGUGGGUUACACCCCAAUUUUACAAUUCCAGGACCGAUCCGAUCCCCAG .............................(((((((.(.(((((((((((.....))))((((((.(((....))).))))))...................))))))).).))))))). (-33.54 = -34.14 + 0.60)

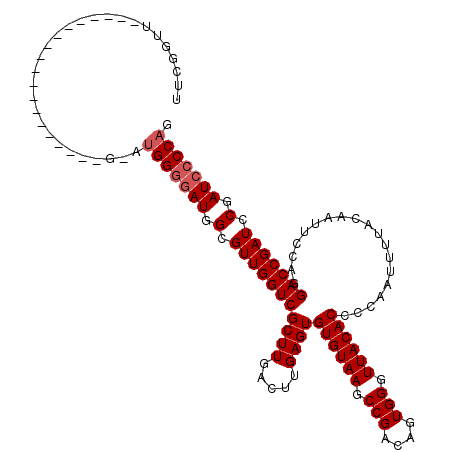
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:35:28 2006