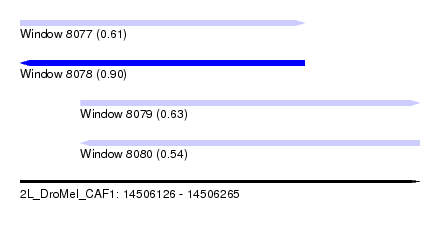
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,506,126 – 14,506,265 |
| Length | 139 |
| Max. P | 0.904488 |

| Location | 14,506,126 – 14,506,225 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 93.33 |
| Mean single sequence MFE | -20.59 |
| Consensus MFE | -15.68 |
| Energy contribution | -16.44 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.606766 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14506126 99 + 22407834 GACAUAUGACUGUCAGACUUUUCUUUUUUUCUUGUCCAAAGAUUAGCCAUUUUGACAUUUUUGUGUCAAAAUAUUUGCGCAUUCAAAUGCUGCGACUAA ((((((.((.(((((((((..(((((...........)))))..))....))))))).)).)))))).......((((((((....)))).)))).... ( -22.60) >DroSec_CAF1 37783 99 + 1 GACAUAUGACUGUCAGACUUUUCUUUUUUUCUUGUCCAAAGAUUAGCCAUUUUGACAUUUUUGUGUCAAAAUAUUUGCGCAUUCAAAUGCUGCGACUAA ((((((.((.(((((((((..(((((...........)))))..))....))))))).)).)))))).......((((((((....)))).)))).... ( -22.60) >DroSim_CAF1 22486 99 + 1 GACAUAUGACUGUCAGACUUCUCUUUUUUUCUUGUCCAAAGAUUAGCCAUUUUGACAUUUUUGUGUCAAAAUAUUUGCGCAUUCAAAUGCUGCGACUAA ((((((.((.(((((((((..(((((...........)))))..))....))))))).)).)))))).......((((((((....)))).)))).... ( -21.80) >DroEre_CAF1 31343 98 + 1 GACGUAUGACUGGCAGCCUCUUCCUU-UUUCAUGUCCAAAGAUUAGCCAUUUUGACAUUUUUGUGUCAAAAUAUUUGCGUAUUCAAAUUCUGCGACUAA .(((((.....(((....((((....-...........))))...)))((((((((((....))))))))))...)))))................... ( -17.76) >DroYak_CAF1 27047 98 + 1 GGCAUAUGACUGUCAGCCGUUUCUUU-UUUCUUGUCCAAAGAUUAGCCACUUUGACAUUUUUGUGUCAAAAUAUUUGCGCAUUCAAACUCUGCGACUAG ((((((.((.((((((..((..((..-..((((.....))))..))..)).)))))).)).))))))..........((((.........))))..... ( -18.20) >consensus GACAUAUGACUGUCAGACUUUUCUUUUUUUCUUGUCCAAAGAUUAGCCAUUUUGACAUUUUUGUGUCAAAAUAUUUGCGCAUUCAAAUGCUGCGACUAA ((((......))))...............((((.....))))((((..((((((((((....))))))))))..((((((((....)))).)))))))) (-15.68 = -16.44 + 0.76)


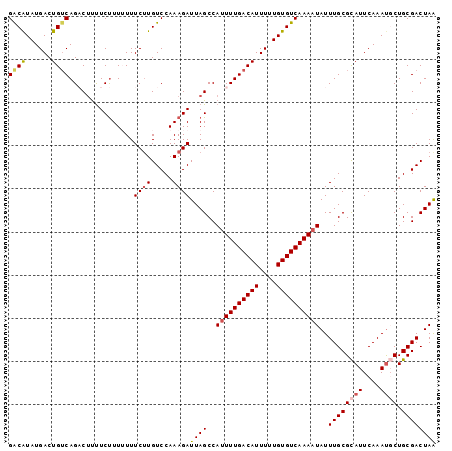
| Location | 14,506,126 – 14,506,225 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 93.33 |
| Mean single sequence MFE | -21.40 |
| Consensus MFE | -17.02 |
| Energy contribution | -17.14 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.904488 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14506126 99 - 22407834 UUAGUCGCAGCAUUUGAAUGCGCAAAUAUUUUGACACAAAAAUGUCAAAAUGGCUAAUCUUUGGACAAGAAAAAAAGAAAAGUCUGACAGUCAUAUGUC ((((((((.((((....))))))....(((((((((......)))))))))))))))(((((...........))))).......((((......)))) ( -22.60) >DroSec_CAF1 37783 99 - 1 UUAGUCGCAGCAUUUGAAUGCGCAAAUAUUUUGACACAAAAAUGUCAAAAUGGCUAAUCUUUGGACAAGAAAAAAAGAAAAGUCUGACAGUCAUAUGUC ((((((((.((((....))))))....(((((((((......)))))))))))))))(((((...........))))).......((((......)))) ( -22.60) >DroSim_CAF1 22486 99 - 1 UUAGUCGCAGCAUUUGAAUGCGCAAAUAUUUUGACACAAAAAUGUCAAAAUGGCUAAUCUUUGGACAAGAAAAAAAGAGAAGUCUGACAGUCAUAUGUC ...(((((.((((....))))))....(((((((((......)))))))))((((..(((((...........)))))..)))).)))........... ( -23.10) >DroEre_CAF1 31343 98 - 1 UUAGUCGCAGAAUUUGAAUACGCAAAUAUUUUGACACAAAAAUGUCAAAAUGGCUAAUCUUUGGACAUGAAA-AAGGAAGAGGCUGCCAGUCAUACGUC ((((((.....(((((......)))))(((((((((......)))))))))))))))(((((..........-)))))...(((.....)))....... ( -18.10) >DroYak_CAF1 27047 98 - 1 CUAGUCGCAGAGUUUGAAUGCGCAAAUAUUUUGACACAAAAAUGUCAAAGUGGCUAAUCUUUGGACAAGAAA-AAAGAAACGGCUGACAGUCAUAUGCC .(((((((...(((((......)))))..(((((((......)))))))))))))).(((((..........-)))))...(((............))) ( -20.60) >consensus UUAGUCGCAGCAUUUGAAUGCGCAAAUAUUUUGACACAAAAAUGUCAAAAUGGCUAAUCUUUGGACAAGAAAAAAAGAAAAGUCUGACAGUCAUAUGUC ((((((((.((((....))))))....(((((((((......)))))))))))))))(((((...........)))))..................... (-17.02 = -17.14 + 0.12)

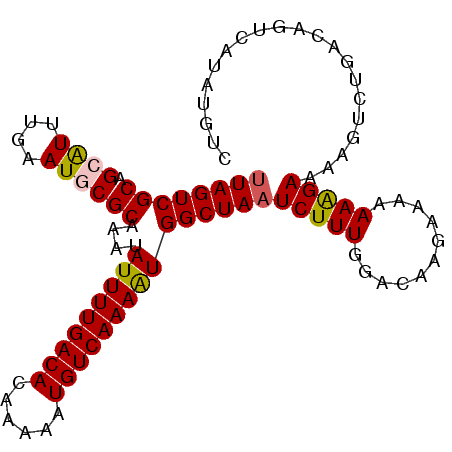
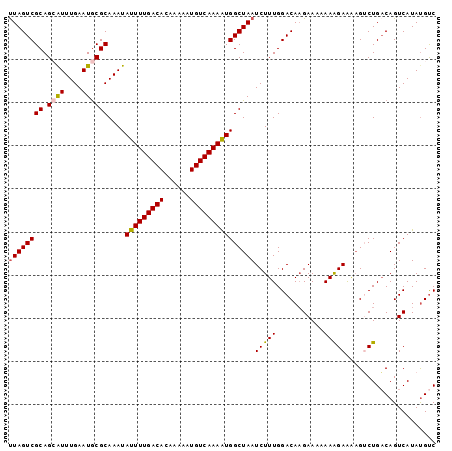
| Location | 14,506,147 – 14,506,265 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 92.40 |
| Mean single sequence MFE | -26.30 |
| Consensus MFE | -21.00 |
| Energy contribution | -22.20 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.634793 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
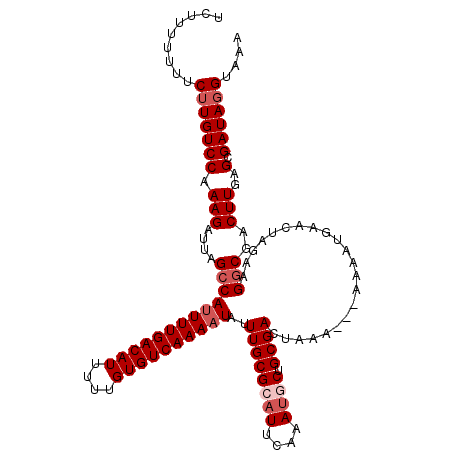
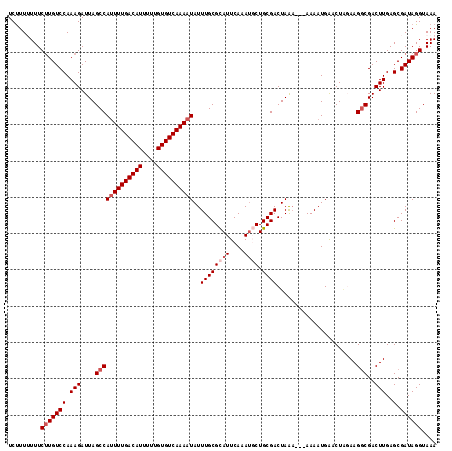
>2L_DroMel_CAF1 14506147 118 + 22407834 UCUUUUUUUCUUGUCCAAAGAUUAGCCAUUUUGACAUUUUUGUGUCAAAAUAUUUGCGCAUUCAAAUGCUGCGACUAAAAAAAAAAUGAACUUUAAGGCGACUUGAGCGAUAGGUAAA .........(((((((.(((....(((((((((((((....))))))))))..((((((((....)))).))))......................)))..)))..).)))))).... ( -26.90) >DroSec_CAF1 37804 116 + 1 UCUUUUUUUCUUGUCCAAAGAUUAGCCAUUUUGACAUUUUUGUGUCAAAAUAUUUGCGCAUUCAAAUGCUGCGACUAAAA--AAAAUGAACUGGAAGGCGACUUGAGCGAUAGGUAAA .........(((((((.(((....(((((((((((((....))))))))))..((((((((....)))).))))......--..............)))..)))..).)))))).... ( -26.90) >DroSim_CAF1 22507 115 + 1 UCUUUUUUUCUUGUCCAAAGAUUAGCCAUUUUGACAUUUUUGUGUCAAAAUAUUUGCGCAUUCAAAUGCUGCGACUAAA---AAAAUGAACUGGAAGGCGACUUGAGCGAUAGGUAAA .........(((((((.(((....(((((((((((((....))))))))))..((((((((....)))).)))).....---..............)))..)))..).)))))).... ( -26.90) >DroEre_CAF1 31364 116 + 1 UCCUU-UUUCAUGUCCAAAGAUUAGCCAUUUUGACAUUUUUGUGUCAAAAUAUUUGCGUAUUCAAAUUCUGCGACUAAAC-AAAAAUGAGCUACAAGGCGACUUGAGCGAUAGGUAAA .(((.-..((..(((...(((......((((((((((....))))))))))(((((......))))))))..))).....-........(((.((((....))))))))).))).... ( -26.30) >DroYak_CAF1 27068 114 + 1 UCUUU-UUUCUUGUCCAAAGAUUAGCCACUUUGACAUUUUUGUGUCAAAAUAUUUGCGCAUUCAAACUCUGCGACUAG---GAAAAUGGGCUAGAAGCCGACUUGAGCGAUAGGUAAA .....-...((((((......((((((..((((((((....)))))))).......((((.........)))).....---.......))))))..((........)))))))).... ( -24.50) >consensus UCUUUUUUUCUUGUCCAAAGAUUAGCCAUUUUGACAUUUUUGUGUCAAAAUAUUUGCGCAUUCAAAUGCUGCGACUAAA___AAAAUGAACUAGAAGGCGACUUGAGCGAUAGGUAAA .........(((((((.(((....(((((((((((((....))))))))))..((((((((....)))).))))......................)))..)))..).)))))).... (-21.00 = -22.20 + 1.20)

| Location | 14,506,147 – 14,506,265 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 92.40 |
| Mean single sequence MFE | -24.60 |
| Consensus MFE | -18.64 |
| Energy contribution | -19.16 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.539489 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
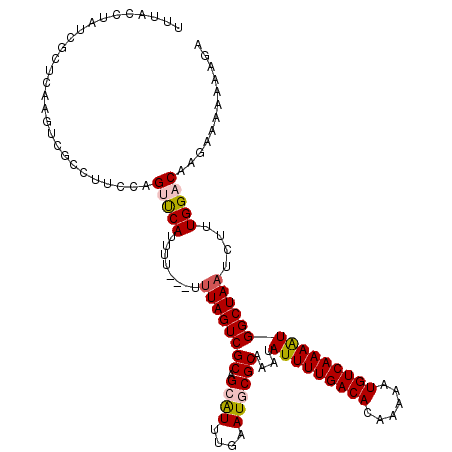
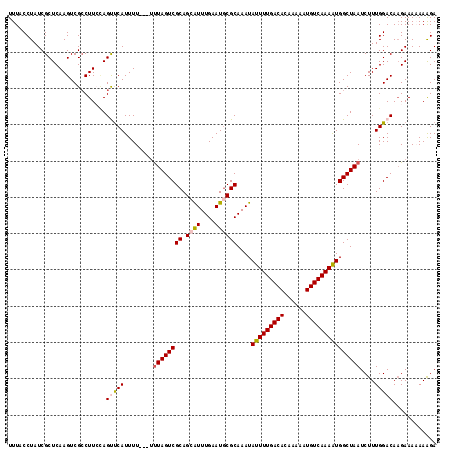
>2L_DroMel_CAF1 14506147 118 - 22407834 UUUACCUAUCGCUCAAGUCGCCUUAAAGUUCAUUUUUUUUUUAGUCGCAGCAUUUGAAUGCGCAAAUAUUUUGACACAAAAAUGUCAAAAUGGCUAAUCUUUGGACAAGAAAAAAAGA ................................((((((((((.(((((.((((....))))))...((((((((((......))))))))))...........))).)))))))))). ( -23.30) >DroSec_CAF1 37804 116 - 1 UUUACCUAUCGCUCAAGUCGCCUUCCAGUUCAUUUU--UUUUAGUCGCAGCAUUUGAAUGCGCAAAUAUUUUGACACAAAAAUGUCAAAAUGGCUAAUCUUUGGACAAGAAAAAAAGA ......................(((..(((((....--..((((((((.((((....))))))....(((((((((......)))))))))))))))....)))))..)))....... ( -24.50) >DroSim_CAF1 22507 115 - 1 UUUACCUAUCGCUCAAGUCGCCUUCCAGUUCAUUUU---UUUAGUCGCAGCAUUUGAAUGCGCAAAUAUUUUGACACAAAAAUGUCAAAAUGGCUAAUCUUUGGACAAGAAAAAAAGA ......................(((..(((((....---.((((((((.((((....))))))....(((((((((......)))))))))))))))....)))))..)))....... ( -25.10) >DroEre_CAF1 31364 116 - 1 UUUACCUAUCGCUCAAGUCGCCUUGUAGCUCAUUUUU-GUUUAGUCGCAGAAUUUGAAUACGCAAAUAUUUUGACACAAAAAUGUCAAAAUGGCUAAUCUUUGGACAUGAAA-AAGGA ....(((...(((((((....)))).)))((((.(..-(.((((((.....(((((......)))))(((((((((......)))))))))))))))....)..).))))..-.))). ( -26.10) >DroYak_CAF1 27068 114 - 1 UUUACCUAUCGCUCAAGUCGGCUUCUAGCCCAUUUUC---CUAGUCGCAGAGUUUGAAUGCGCAAAUAUUUUGACACAAAAAUGUCAAAGUGGCUAAUCUUUGGACAAGAAA-AAAGA ................((((((.....))).......---.(((((((...(((((......)))))..(((((((......)))))))))))))).......)))......-..... ( -24.00) >consensus UUUACCUAUCGCUCAAGUCGCCUUCCAGUUCAUUUU___UUUAGUCGCAGCAUUUGAAUGCGCAAAUAUUUUGACACAAAAAUGUCAAAAUGGCUAAUCUUUGGACAAGAAAAAAAGA ...........................(((((........((((((((.((((....))))))....(((((((((......)))))))))))))))....)))))............ (-18.64 = -19.16 + 0.52)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:35:21 2006