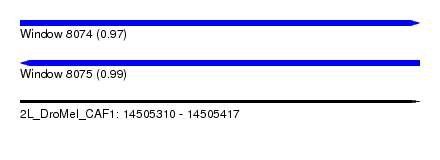
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,505,310 – 14,505,417 |
| Length | 107 |
| Max. P | 0.991946 |

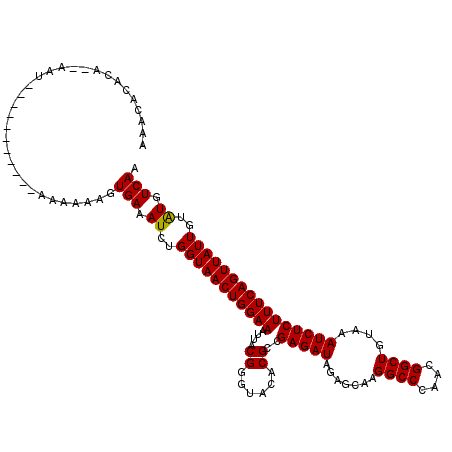
| Location | 14,505,310 – 14,505,417 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.53 |
| Mean single sequence MFE | -28.66 |
| Consensus MFE | -22.08 |
| Energy contribution | -22.52 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.58 |
| SVM RNA-class probability | 0.965359 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14505310 107 + 22407834 UUGACAUACAAUAACUGAAAGAGAUUUACAGCCGUUGGGCCUUGCUCUAUCUCGGCGUGUACCCGUAAUUUCCAGUUACCAGAUUUCACUUUUUU-----------AUU--UGUGUGUUU ..(((((((((....((((((((...((((((((..((((...)))).....)))).))))...(((((.....))))).........)))))))-----------).)--)))))))). ( -28.20) >DroSec_CAF1 36984 107 + 1 UUGACAUACAAUAACUGAAAGAGAUUUACAGCCGUUGGGCCUCGCUCUAUCUCGGCGUGUACCCGUAAUUUCCAGUUACCAGAUUUCACUUUUUU-----------AUU--UGUGUGUUU ..(((((((((....((((((((...((((((((..((((...)))).....)))).))))...(((((.....))))).........)))))))-----------).)--)))))))). ( -28.20) >DroSim_CAF1 21690 107 + 1 UUGACAUACAAUAACUGAAAGAGAUUUACAGCCGUUGGGCCUCGCUCUAUCUCGGCGUGUACCCGUAAUUUCCAGUUACCAGAUUUCACUUUUUU-----------AUU--UGUGUGUUU ..(((((((((....((((((((...((((((((..((((...)))).....)))).))))...(((((.....))))).........)))))))-----------).)--)))))))). ( -28.20) >DroEre_CAF1 30561 119 + 1 UUGACACACAAUAACUGAAAGAGAUUUACAGCCAG-GGGCCUUGCUCUAUCUCGGCGUGUAGCCGUAAUUUCCAGUUACCAGAUUUCACCUUUUUUUCUGGUUUUUAUUUCUGUGUGUUU ..((((((((.((((((...(((((....(((.((-....)).)))..)))))(((.....)))........))))))(((((.............)))))..........)))))))). ( -36.12) >DroYak_CAF1 26237 106 + 1 UUGACAUACAAUAACUGAAAGAGAUUUACAGCCCGUGGGCCUUGCUCUAUCUCGUCGUGUACCCGUAAUUUCCAGUUACCAGAUUUCACUUUUU------------AUU--UGUGUGUUU ..(((((((((....((((((((((...(((((....)))..))....)))))...(((.....(((((.....))))).......))).))))------------).)--)))))))). ( -22.60) >consensus UUGACAUACAAUAACUGAAAGAGAUUUACAGCCGUUGGGCCUUGCUCUAUCUCGGCGUGUACCCGUAAUUUCCAGUUACCAGAUUUCACUUUUUU___________AUU__UGUGUGUUU ..((((((((.((((((...((((((((((((((..((((...)))).....)))).)))).....))))))))))))...(....)........................)))))))). (-22.08 = -22.52 + 0.44)

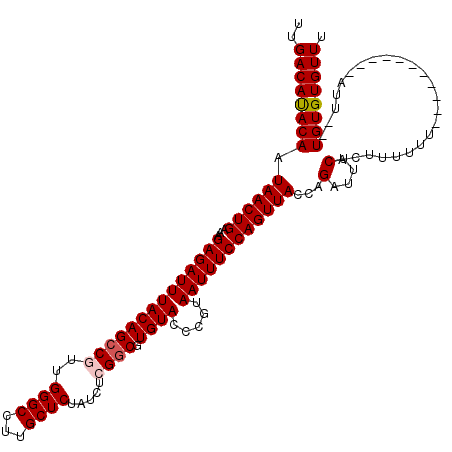
| Location | 14,505,310 – 14,505,417 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.53 |
| Mean single sequence MFE | -26.74 |
| Consensus MFE | -22.74 |
| Energy contribution | -22.58 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.85 |
| SVM decision value | 2.30 |
| SVM RNA-class probability | 0.991946 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14505310 107 - 22407834 AAACACACA--AAU-----------AAAAAAGUGAAAUCUGGUAACUGGAAAUUACGGGUACACGCCGAGAUAGAGCAAGGCCCAACGGCUGUAAAUCUCUUUCAGUUAUUGUAUGUCAA .........--...-----------.......(((.((..(((((((((((......(((....)))(((((.......((((....))))....))))))))))))))))..)).))). ( -23.90) >DroSec_CAF1 36984 107 - 1 AAACACACA--AAU-----------AAAAAAGUGAAAUCUGGUAACUGGAAAUUACGGGUACACGCCGAGAUAGAGCGAGGCCCAACGGCUGUAAAUCUCUUUCAGUUAUUGUAUGUCAA .........--...-----------.......(((.((..(((((((((((......(((....)))(((((.......((((....))))....))))))))))))))))..)).))). ( -23.90) >DroSim_CAF1 21690 107 - 1 AAACACACA--AAU-----------AAAAAAGUGAAAUCUGGUAACUGGAAAUUACGGGUACACGCCGAGAUAGAGCGAGGCCCAACGGCUGUAAAUCUCUUUCAGUUAUUGUAUGUCAA .........--...-----------.......(((.((..(((((((((((......(((....)))(((((.......((((....))))....))))))))))))))))..)).))). ( -23.90) >DroEre_CAF1 30561 119 - 1 AAACACACAGAAAUAAAAACCAGAAAAAAAGGUGAAAUCUGGUAACUGGAAAUUACGGCUACACGCCGAGAUAGAGCAAGGCCC-CUGGCUGUAAAUCUCUUUCAGUUAUUGUGUGUCAA ..((((((((........((((((.............))))))((((((((.....(((.....)))((((((.(((.((....-)).))).))..)))))))))))).))))))))... ( -37.32) >DroYak_CAF1 26237 106 - 1 AAACACACA--AAU------------AAAAAGUGAAAUCUGGUAACUGGAAAUUACGGGUACACGACGAGAUAGAGCAAGGCCCACGGGCUGUAAAUCUCUUUCAGUUAUUGUAUGUCAA .........--...------------......(((.((..(((((((((((....((......))..(((((.......((((....))))....))))))))))))))))..)).))). ( -24.70) >consensus AAACACACA__AAU___________AAAAAAGUGAAAUCUGGUAACUGGAAAUUACGGGUACACGCCGAGAUAGAGCAAGGCCCAACGGCUGUAAAUCUCUUUCAGUUAUUGUAUGUCAA ................................(((.((..(((((((((((....((......))..(((((.......((((....))))....))))))))))))))))..)).))). (-22.74 = -22.58 + -0.16)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:35:17 2006