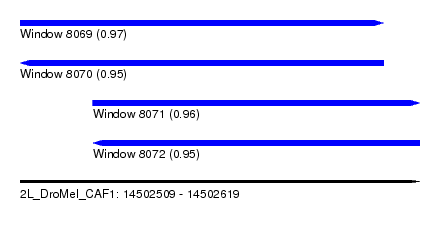
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,502,509 – 14,502,619 |
| Length | 110 |
| Max. P | 0.965536 |

| Location | 14,502,509 – 14,502,609 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.74 |
| Mean single sequence MFE | -37.85 |
| Consensus MFE | -31.51 |
| Energy contribution | -31.52 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.09 |
| Mean z-score | -3.17 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.58 |
| SVM RNA-class probability | 0.965536 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
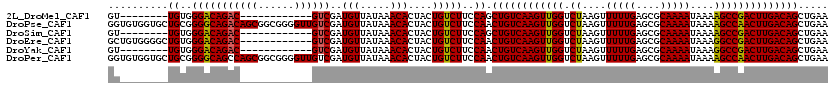
>2L_DroMel_CAF1 14502509 100 + 22407834 GU--------UGUGGGACAGAC------------GUCGAUGUUAUAAACACUACUGUCUUCCAGCUGUCAAGUUGGUCUAAGUUUUUGAGCGCAAAAUAAAAGCCGACUUGACAGCUGAA ..--------....(((.((((------------((.(.(((.....)))).)).)))))))(((((((((((((((......(((((....))))).....)))))))))))))))... ( -35.60) >DroPse_CAF1 23226 120 + 1 GGUGUGGUGCUGCGGGGCAGACAGCGGCGGGGUUGUCGAUGUUAUAAACACUACUGUCUUCCAACUGUCAAGUUGGUCUAAGUUUUUGAGCGCAAAAUAAAAGCCAACUUGACAGCUGAA ...((((((......((((((((((......))))))..)))).....)))))).......((.(((((((((((((......(((((....))))).....))))))))))))).)).. ( -39.90) >DroSim_CAF1 18558 100 + 1 GU--------UGUGGGACAGAC------------GUCGAUGUUAUAAACACUACUGUCUUCCAGCUGUCAAGUUGGUCUAAGUUUUUGAGCGCAAAAUAAAAGCCGACUUGACAGCUGAA ..--------....(((.((((------------((.(.(((.....)))).)).)))))))(((((((((((((((......(((((....))))).....)))))))))))))))... ( -35.60) >DroEre_CAF1 27920 108 + 1 GCUGUGGGGCUGUGGGACAGAC------------GUCGAUGUUAUAAACACUACUGUCUUCCAACUGUCAAGUUGGUCUAAGUUUUUGAGCGCAAAAUAAAGGCCGACUUGACAGCUGAA ..((..((((.(((((((....------------)))..(((.....))))))).))))..)).(((((((((((((((....(((((....)))))...)))))))))))))))..... ( -39.30) >DroYak_CAF1 23369 100 + 1 GU--------UGUGGGACAGAC------------GUCGAUGUUAUAAACACUACUGUCUUCCAACUGUCAAGUUGGUCUAAGUUUUUGAGCGCAAAAUAAAGGCCGACUUGACAGCUGAA ((--------((..((((((..------------...................))))))..))))((((((((((((((....(((((....)))))...))))))))))))))...... ( -35.60) >DroPer_CAF1 22725 120 + 1 GGUGUGGUGCUGCGGGGCAGCCAGCGGCGGGGUUGUCGAUGUUAUAAACACUACUGUCUUCCAACUGUCAAGUUGGUCUAAGUUUUUGAGCGCAAAAUAAAAGCCAACUUGACAGCUGAA ...((((((..(((.(((((((........)))))))..)))......)))))).......((.(((((((((((((......(((((....))))).....))))))))))))).)).. ( -41.10) >consensus GU________UGUGGGACAGAC____________GUCGAUGUUAUAAACACUACUGUCUUCCAACUGUCAAGUUGGUCUAAGUUUUUGAGCGCAAAAUAAAAGCCGACUUGACAGCUGAA ..........((..(((((((((((......))))))..(((.....)))....)))))..)).((((((((((((.((....(((((....)))))....))))))))))))))..... (-31.51 = -31.52 + 0.00)
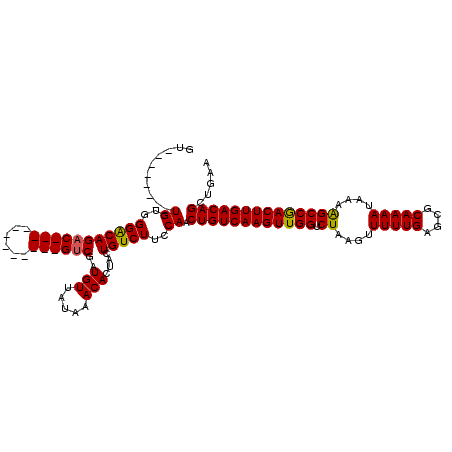
| Location | 14,502,509 – 14,502,609 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.74 |
| Mean single sequence MFE | -35.02 |
| Consensus MFE | -26.89 |
| Energy contribution | -27.33 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.92 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.35 |
| SVM RNA-class probability | 0.945272 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14502509 100 - 22407834 UUCAGCUGUCAAGUCGGCUUUUAUUUUGCGCUCAAAAACUUAGACCAACUUGACAGCUGGAAGACAGUAGUGUUUAUAACAUCGAC------------GUCUGUCCCACA--------AC ((((((((((((((.((((....(((((....)))))....)).)).)))))))))))))).(((((..((((.....))))....------------..))))).....--------.. ( -33.30) >DroPse_CAF1 23226 120 - 1 UUCAGCUGUCAAGUUGGCUUUUAUUUUGCGCUCAAAAACUUAGACCAACUUGACAGUUGGAAGACAGUAGUGUUUAUAACAUCGACAACCCCGCCGCUGUCUGCCCCGCAGCACCACACC (((((((((((((((((((....(((((....)))))....)).)))))))))))))))))((((((..(((...................)))..))))))((......))........ ( -40.81) >DroSim_CAF1 18558 100 - 1 UUCAGCUGUCAAGUCGGCUUUUAUUUUGCGCUCAAAAACUUAGACCAACUUGACAGCUGGAAGACAGUAGUGUUUAUAACAUCGAC------------GUCUGUCCCACA--------AC ((((((((((((((.((((....(((((....)))))....)).)).)))))))))))))).(((((..((((.....))))....------------..))))).....--------.. ( -33.30) >DroEre_CAF1 27920 108 - 1 UUCAGCUGUCAAGUCGGCCUUUAUUUUGCGCUCAAAAACUUAGACCAACUUGACAGUUGGAAGACAGUAGUGUUUAUAACAUCGAC------------GUCUGUCCCACAGCCCCACAGC ((((((((((((((.((.((...(((((....)))))....)).)).)))))))))))))).(((((..((((.....))))....------------..)))))............... ( -30.50) >DroYak_CAF1 23369 100 - 1 UUCAGCUGUCAAGUCGGCCUUUAUUUUGCGCUCAAAAACUUAGACCAACUUGACAGUUGGAAGACAGUAGUGUUUAUAACAUCGAC------------GUCUGUCCCACA--------AC ((((((((((((((.((.((...(((((....)))))....)).)).)))))))))))))).(((((..((((.....))))....------------..))))).....--------.. ( -30.50) >DroPer_CAF1 22725 120 - 1 UUCAGCUGUCAAGUUGGCUUUUAUUUUGCGCUCAAAAACUUAGACCAACUUGACAGUUGGAAGACAGUAGUGUUUAUAACAUCGACAACCCCGCCGCUGGCUGCCCCGCAGCACCACACC (((((((((((((((((((....(((((....)))))....)).)))))))))))))))))...(((..(((...................)))..)))(((((...)))))........ ( -41.71) >consensus UUCAGCUGUCAAGUCGGCUUUUAUUUUGCGCUCAAAAACUUAGACCAACUUGACAGUUGGAAGACAGUAGUGUUUAUAACAUCGAC____________GUCUGUCCCACA________AC ((((((((((((((.((((....(((((....)))))....)).)).)))))))))))))).(((((..((((.....))))..................)))))............... (-26.89 = -27.33 + 0.44)
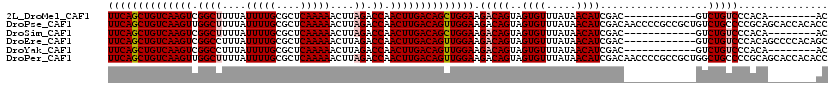
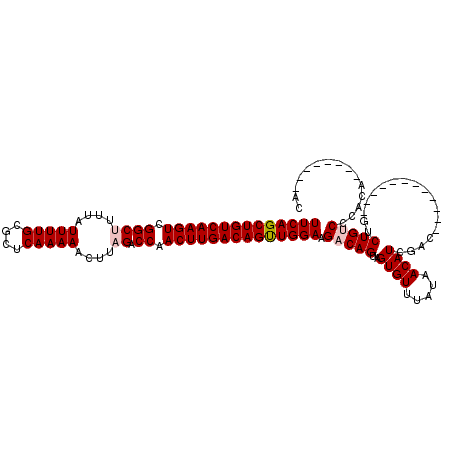
| Location | 14,502,529 – 14,502,619 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.73 |
| Mean single sequence MFE | -32.67 |
| Consensus MFE | -26.54 |
| Energy contribution | -26.10 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.58 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.57 |
| SVM RNA-class probability | 0.964839 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14502529 90 + 22407834 GUUAUAAACACUACUGUCUUCCAGCUGUCAAGUUGGUCUAAGUUUUUGAGCGCAAAAUAAAAGCCGACUUGACAGCUGAACGGUCGACCA------------------------------ ..........(.(((((....((((((((((((((((......(((((....))))).....)))))))))))))))).))))).)....------------------------------ ( -33.30) >DroPse_CAF1 23266 120 + 1 GUUAUAAACACUACUGUCUUCCAACUGUCAAGUUGGUCUAAGUUUUUGAGCGCAAAAUAAAAGCCAACUUGACAGCUGAACGGUCUGUCUCCCUUCCGUCUGUUGGCGGCCAAACUGAUG ............(((((....((.(((((((((((((......(((((....))))).....))))))))))))).)).)))))..(((......(((((....))))).......))). ( -34.02) >DroSim_CAF1 18578 90 + 1 GUUAUAAACACUACUGUCUUCCAGCUGUCAAGUUGGUCUAAGUUUUUGAGCGCAAAAUAAAAGCCGACUUGACAGCUGAACGGUCGACCA------------------------------ ..........(.(((((....((((((((((((((((......(((((....))))).....)))))))))))))))).))))).)....------------------------------ ( -33.30) >DroEre_CAF1 27948 90 + 1 GUUAUAAACACUACUGUCUUCCAACUGUCAAGUUGGUCUAAGUUUUUGAGCGCAAAAUAAAGGCCGACUUGACAGCUGAACGGUCGACCA------------------------------ ..........(.(((((....((.(((((((((((((((....(((((....)))))...))))))))))))))).)).))))).)....------------------------------ ( -30.70) >DroYak_CAF1 23389 90 + 1 GUUAUAAACACUACUGUCUUCCAACUGUCAAGUUGGUCUAAGUUUUUGAGCGCAAAAUAAAGGCCGACUUGACAGCUGAACGGUCGACCA------------------------------ ..........(.(((((....((.(((((((((((((((....(((((....)))))...))))))))))))))).)).))))).)....------------------------------ ( -30.70) >DroPer_CAF1 22765 120 + 1 GUUAUAAACACUACUGUCUUCCAACUGUCAAGUUGGUCUAAGUUUUUGAGCGCAAAAUAAAAGCCAACUUGACAGCUGAACGGUCUGUCUCCCUUCCGUCUGUUGGCGGCCAAACUGAUG ............(((((....((.(((((((((((((......(((((....))))).....))))))))))))).)).)))))..(((......(((((....))))).......))). ( -34.02) >consensus GUUAUAAACACUACUGUCUUCCAACUGUCAAGUUGGUCUAAGUUUUUGAGCGCAAAAUAAAAGCCGACUUGACAGCUGAACGGUCGACCA______________________________ ............(((((....((.((((((((((((.((....(((((....)))))....)))))))))))))).)).))))).................................... (-26.54 = -26.10 + -0.44)


| Location | 14,502,529 – 14,502,619 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.73 |
| Mean single sequence MFE | -32.90 |
| Consensus MFE | -26.42 |
| Energy contribution | -26.53 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.14 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.45 |
| SVM RNA-class probability | 0.954954 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14502529 90 - 22407834 ------------------------------UGGUCGACCGUUCAGCUGUCAAGUCGGCUUUUAUUUUGCGCUCAAAAACUUAGACCAACUUGACAGCUGGAAGACAGUAGUGUUUAUAAC ------------------------------..(((.....((((((((((((((.((((....(((((....)))))....)).)).)))))))))))))).)))............... ( -30.90) >DroPse_CAF1 23266 120 - 1 CAUCAGUUUGGCCGCCAACAGACGGAAGGGAGACAGACCGUUCAGCUGUCAAGUUGGCUUUUAUUUUGCGCUCAAAAACUUAGACCAACUUGACAGUUGGAAGACAGUAGUGUUUAUAAC .....(((((........)))))((...(....)...)).(((((((((((((((((((....(((((....)))))....)).)))))))))))))))))(((((....)))))..... ( -39.70) >DroSim_CAF1 18578 90 - 1 ------------------------------UGGUCGACCGUUCAGCUGUCAAGUCGGCUUUUAUUUUGCGCUCAAAAACUUAGACCAACUUGACAGCUGGAAGACAGUAGUGUUUAUAAC ------------------------------..(((.....((((((((((((((.((((....(((((....)))))....)).)).)))))))))))))).)))............... ( -30.90) >DroEre_CAF1 27948 90 - 1 ------------------------------UGGUCGACCGUUCAGCUGUCAAGUCGGCCUUUAUUUUGCGCUCAAAAACUUAGACCAACUUGACAGUUGGAAGACAGUAGUGUUUAUAAC ------------------------------..(((.....((((((((((((((.((.((...(((((....)))))....)).)).)))))))))))))).)))............... ( -28.10) >DroYak_CAF1 23389 90 - 1 ------------------------------UGGUCGACCGUUCAGCUGUCAAGUCGGCCUUUAUUUUGCGCUCAAAAACUUAGACCAACUUGACAGUUGGAAGACAGUAGUGUUUAUAAC ------------------------------..(((.....((((((((((((((.((.((...(((((....)))))....)).)).)))))))))))))).)))............... ( -28.10) >DroPer_CAF1 22765 120 - 1 CAUCAGUUUGGCCGCCAACAGACGGAAGGGAGACAGACCGUUCAGCUGUCAAGUUGGCUUUUAUUUUGCGCUCAAAAACUUAGACCAACUUGACAGUUGGAAGACAGUAGUGUUUAUAAC .....(((((........)))))((...(....)...)).(((((((((((((((((((....(((((....)))))....)).)))))))))))))))))(((((....)))))..... ( -39.70) >consensus ______________________________UGGUCGACCGUUCAGCUGUCAAGUCGGCUUUUAUUUUGCGCUCAAAAACUUAGACCAACUUGACAGUUGGAAGACAGUAGUGUUUAUAAC ........................................((((((((((((((.((((....(((((....)))))....)).)).))))))))))))))(((((....)))))..... (-26.42 = -26.53 + 0.11)

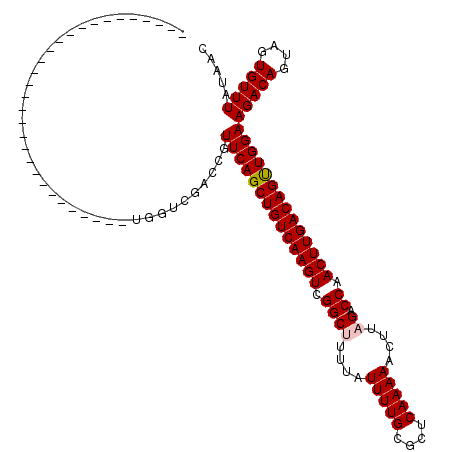
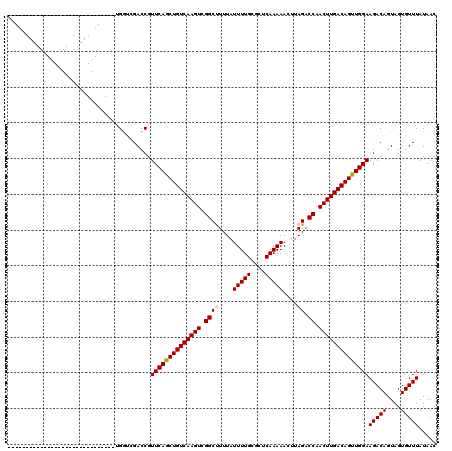
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:35:14 2006