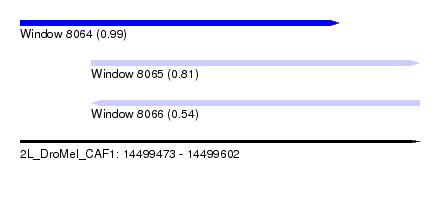
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,499,473 – 14,499,602 |
| Length | 129 |
| Max. P | 0.994362 |

| Location | 14,499,473 – 14,499,576 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 96.00 |
| Mean single sequence MFE | -23.41 |
| Consensus MFE | -21.50 |
| Energy contribution | -21.54 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.47 |
| SVM RNA-class probability | 0.994362 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14499473 103 + 22407834 -----GCCGCUGUCCGAUGGCCACAUUAUUAAGCCAAUUCAGCGGCAGCAGAUAAAUAAAUAAAAUUUGAAACAUUGCCGCAACAAUAAAAGGCGACAGUACACGAUU -----(((((((.....((((...........))))...)))))))..(((((...........))))).....(((((............)))))............ ( -23.80) >DroSec_CAF1 31178 103 + 1 -----GCCGCUGUCCGAUGGCCACAUUAUUAAGCCAAUUCAGCGGCAGCAGAUAAAUAAAUAAAAUUUGAAACAUUGCCGCAACAAUAAAAGGCGACAGUACACGAUU -----(((((((.....((((...........))))...)))))))..(((((...........))))).....(((((............)))))............ ( -23.80) >DroSim_CAF1 15212 103 + 1 -----GCCGCUGUCCGAUGGCCACAUUAUUAAGCCAAUUCAGCGGCAGCAGAUAAAUAAAUAAAAUUUGAAACAUUGCCGCAACAAUAAAAGGCGACAGUACACGAUU -----(((((((.....((((...........))))...)))))))..(((((...........))))).....(((((............)))))............ ( -23.80) >DroYak_CAF1 20434 108 + 1 CUCUUGCUGCUGUCCGAUGGCCACAUUAUUAAGCCAAUUCAGCGGCAGCAGAUAAAUAAAUAAAAUUUGAAACAUUGCCGCAACAAUAAAAGGCGACAGUACACGAUU .((.((.((((((((..((((...........)))).....((((((((((((...........))))).....)))))))...........).))))))))).)).. ( -26.30) >DroPer_CAF1 17943 103 + 1 -----UGUGUUGUCCGAUGGCCACAUUAUUAAGCCAAUUCAACGGCAGCAGAUAAAUAAAUAAAAUUUGAAACAUUGCCGCAACAAUAAAAGGCGACAGUACACGAUU -----(((((((((.((((....)))).....(((.......(((((((((((...........))))).....))))))...........))))))))))))..... ( -19.37) >consensus _____GCCGCUGUCCGAUGGCCACAUUAUUAAGCCAAUUCAGCGGCAGCAGAUAAAUAAAUAAAAUUUGAAACAUUGCCGCAACAAUAAAAGGCGACAGUACACGAUU ........(((((((..((((...........)))).....((((((((((((...........))))).....)))))))...........).))))))........ (-21.50 = -21.54 + 0.04)

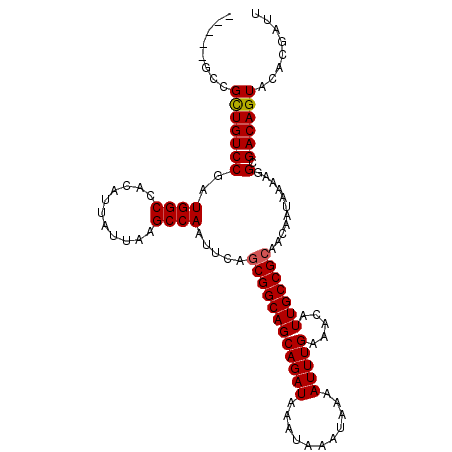
| Location | 14,499,496 – 14,499,602 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.13 |
| Mean single sequence MFE | -27.13 |
| Consensus MFE | -19.28 |
| Energy contribution | -19.76 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.812436 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14499496 106 + 22407834 UUAAGCCAAUUCAGCGGCAGCAGAUAAAUAAAUAAAAUUUGAAACAUUGCCGCAACAAUAAAAGGCGACAGUACACGAUUUAUUAAACGGAGUGGCUGCUG------------C--UGGG ..........(((((((((((...(((((.......)))))...((((.(((....((((((...((........)).))))))...))))))))))))))------------)--))). ( -27.80) >DroSec_CAF1 31201 112 + 1 UUAAGCCAAUUCAGCGGCAGCAGAUAAAUAAAUAAAAUUUGAAACAUUGCCGCAACAAUAAAAGGCGACAGUACACGAUUUAUUAAACGGAGUGGCUGCUG------CUGCUGC--CGGG .....((....((((((((((((.(((((.......)))))...((((.(((....((((((...((........)).))))))...))))))).))))))------)))))).--..)) ( -31.50) >DroEre_CAF1 25171 108 + 1 UUAAGCCAAUUCAGCGGCAGCAGAUAAAUAAAUAAAAUUUGAAACAUUGCCGCAACAAUAAAAGGCGACAGUACACGAUUUAUUAAACGGAGUGGCUGCUGCUG------------CUGU ...........((((((((((((.(((((.......)))))...((((.(((....((((((...((........)).))))))...))))))).)))))))))------------))). ( -30.30) >DroYak_CAF1 20462 120 + 1 UUAAGCCAAUUCAGCGGCAGCAGAUAAAUAAAUAAAAUUUGAAACAUUGCCGCAACAAUAAAAGGCGACAGUACACGAUUUAUUAAACGGAGUGGCUGCUGCUGCUUCUGCUUCUGCUGG ...........((((((.((((((..(((((((.....(((.....(((((............))))))))......)))))))......((..((....))..)))))))).)))))). ( -32.60) >DroPer_CAF1 17966 96 + 1 UUAAGCCAAUUCAACGGCAGCAGAUAAAUAAAUAAAAUUUGAAACAUUGCCGCAACAAUAAAAGGCGACAGUACACGAUUUAUUAAACGAAGCUGU------------------------ ....(((........))).((((.......((((((.(.((.....(((((............))))).....)).).))))))........))))------------------------ ( -13.46) >consensus UUAAGCCAAUUCAGCGGCAGCAGAUAAAUAAAUAAAAUUUGAAACAUUGCCGCAACAAUAAAAGGCGACAGUACACGAUUUAUUAAACGGAGUGGCUGCUG____________C__CGGG ...(((((.(((.((((((((((((...........))))).....)))))))...((((((...((........)).))))))....))).)))))....................... (-19.28 = -19.76 + 0.48)

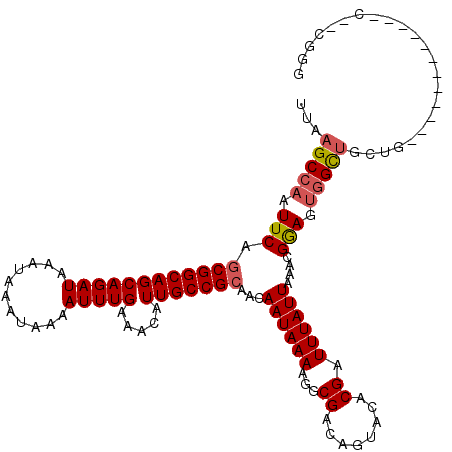
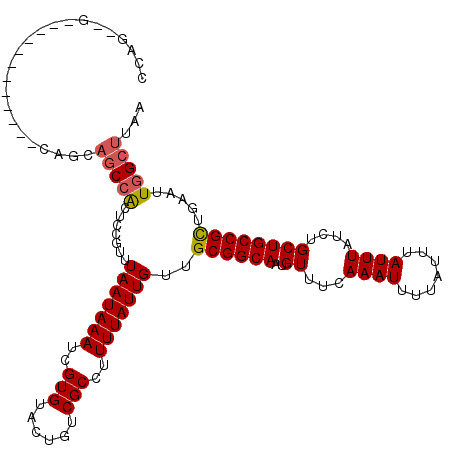
| Location | 14,499,496 – 14,499,602 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.13 |
| Mean single sequence MFE | -26.34 |
| Consensus MFE | -18.10 |
| Energy contribution | -18.38 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.537824 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14499496 106 - 22407834 CCCA--G------------CAGCAGCCACUCCGUUUAAUAAAUCGUGUACUGUCGCCUUUUAUUGUUGCGGCAAUGUUUCAAAUUUUAUUUAUUUAUCUGCUGCCGCUGAAUUGGCUUAA ..((--(------------(.(((((....((((.(((((((..(((......)))..)))))))..)))).........((((.......))))....))))).))))........... ( -25.10) >DroSec_CAF1 31201 112 - 1 CCCG--GCAGCAG------CAGCAGCCACUCCGUUUAAUAAAUCGUGUACUGUCGCCUUUUAUUGUUGCGGCAAUGUUUCAAAUUUUAUUUAUUUAUCUGCUGCCGCUGAAUUGGCUUAA .(((--.((((.(------((((((.....((((.(((((((..(((......)))..)))))))..)))).........((((.......))))..))))))).))))...)))..... ( -30.70) >DroEre_CAF1 25171 108 - 1 ACAG------------CAGCAGCAGCCACUCCGUUUAAUAAAUCGUGUACUGUCGCCUUUUAUUGUUGCGGCAAUGUUUCAAAUUUUAUUUAUUUAUCUGCUGCCGCUGAAUUGGCUUAA .(((------------(.(((((((.....((((.(((((((..(((......)))..)))))))..)))).........((((.......))))..))))))).))))........... ( -29.20) >DroYak_CAF1 20462 120 - 1 CCAGCAGAAGCAGAAGCAGCAGCAGCCACUCCGUUUAAUAAAUCGUGUACUGUCGCCUUUUAUUGUUGCGGCAAUGUUUCAAAUUUUAUUUAUUUAUCUGCUGCCGCUGAAUUGGCUUAA .......((((.....((((.(((((....((((.(((((((..(((......)))..)))))))..)))).........((((.......))))....))))).)))).....)))).. ( -30.40) >DroPer_CAF1 17966 96 - 1 ------------------------ACAGCUUCGUUUAAUAAAUCGUGUACUGUCGCCUUUUAUUGUUGCGGCAAUGUUUCAAAUUUUAUUUAUUUAUCUGCUGCCGUUGAAUUGGCUUAA ------------------------..(((.......((((((...((.((((((((...........))))))..))..))...)))))).........)))((((......)))).... ( -16.29) >consensus CCAG__G____________CAGCAGCCACUCCGUUUAAUAAAUCGUGUACUGUCGCCUUUUAUUGUUGCGGCAAUGUUUCAAAUUUUAUUUAUUUAUCUGCUGCCGCUGAAUUGGCUUAA .......................(((((.......(((((((..(((......)))..)))))))..((((((..((...((((.......))))....)))))))).....)))))... (-18.10 = -18.38 + 0.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:35:08 2006