
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,498,918 – 14,499,064 |
| Length | 146 |
| Max. P | 0.553606 |

| Location | 14,498,918 – 14,499,026 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.18 |
| Mean single sequence MFE | -35.77 |
| Consensus MFE | -14.90 |
| Energy contribution | -15.22 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.35 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.42 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14498918 108 + 22407834 C---GUGGGCUGAUUGGUAAAAUCAG-----GUCACUGGCAGAAAGCC-GUCAGCGGCAAUCG---CCAUACGCAGAUAACUUAUUUAUCGGAAAUGGCUGUUGACAAAUCCGGUAAUGA .---((((.((((((.....))))))-----.))))((((.((..(((-(....))))..)))---)))................((((((((..((........))..))))))))... ( -36.00) >DroSec_CAF1 30623 111 + 1 C---GUGGGCUGAUUGGUAAAAUCAG-----GUCACUGGCAGAAAGCC-GUCAGCGGCAAUCGCCGCCAUUCGCAGAUAACUCAUUUAUCGGAAAUGGCUGUUGACAAAUCCGGUAAUGA .---((((.((((((.....))))))-----.)))).(((.....)))-(((((((((....)))((((((....(((((.....)))))...)))))).)))))).............. ( -40.80) >DroSim_CAF1 14657 111 + 1 C---GUGGGCUGAUUGGUAAAAUCAG-----GUCACUGGCAGAAAGCC-GUCAGCGGCAAUCGCCGCCAUUCGCAGAUAACUCAUUUAUCGGAAAUGGCUGUUGACAAAUCCGGUAAUGA .---((((.((((((.....))))))-----.)))).(((.....)))-(((((((((....)))((((((....(((((.....)))))...)))))).)))))).............. ( -40.80) >DroEre_CAF1 24673 108 + 1 CG----GGACUGAUUGGUAAAAUCAG-----GUCACUGGCAGAAAGCCAGUCGGCGGCAAUCG---GCACUCGCAGAUAACUCAUUUAUCGCAAAUGCUUGUUGACAAAUCCGGUAAUGA ((----((.((((((.....))))))-----(((((((((.....)))))).)))(.((((.(---(((...((.(((((.....)))))))...)))).)))).)...))))....... ( -34.80) >DroYak_CAF1 19884 111 + 1 CAGCGUGGGCUGAUUGGUAAAAUCAG-----GUCACUGGCAGAAAGCC-GUCAGCGGUAAUCG---CCAUUCGCAGAUAACUCAUUUAUCGGAAAUGCUCGUUGACAAAUCCGGUGAUGA ..((((((.((((((.....))))))-----.))))((((.((..(((-(....))))..)))---)))...)).......(((((.((((((..(((.....).))..))))))))))) ( -37.40) >DroPer_CAF1 16965 100 + 1 A---GUGUGGUG--UGGUGCUGUGGGUUUCACUGAAUGGCAUGGAAAU-GUCACCAACAAU------------CAGGCAACACA--UAUCGGAAAUUGUCGCUGACAGAGCUGUUAAUGU .---((((.((.--((((....(((..(((...)))((((((....))-)))))))...))------------)).)).)))).--((.(((...(((((...)))))..))).)).... ( -24.80) >consensus C___GUGGGCUGAUUGGUAAAAUCAG_____GUCACUGGCAGAAAGCC_GUCAGCGGCAAUCG___CCAUUCGCAGAUAACUCAUUUAUCGGAAAUGGCUGUUGACAAAUCCGGUAAUGA ........((((((.(((....((((.........))))......))).))))))..........................((((.(((((((..((........))..))))))))))) (-14.90 = -15.22 + 0.31)

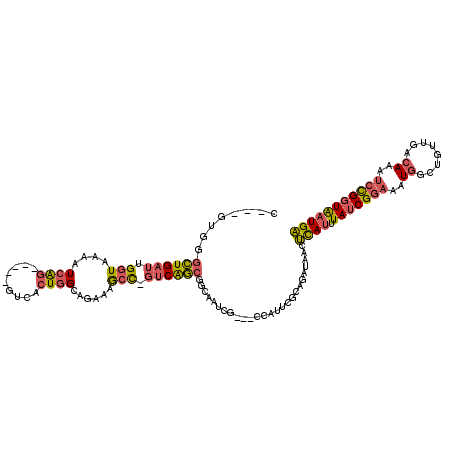

| Location | 14,498,950 – 14,499,064 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.64 |
| Mean single sequence MFE | -29.70 |
| Consensus MFE | -13.84 |
| Energy contribution | -15.07 |
| Covariance contribution | 1.23 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.47 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.553606 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14498950 114 - 22407834 UUUAACGAAUCAUCGAUUAAACGCGAGAUCGUAGA--AAUUCAUUACCGGAUUUGUCAACAGCCAUUUCCGAUAAAUAAGUUAUCUGCGUAUGG---CGAUUGCCGCUGAC-GGCUUUCU ((((((((....))).))))).((.(...((((((--((((..(((.((((..((.(....).))..)))).)))...)))).))))))..).)---)((..((((....)-)))..)). ( -28.40) >DroSec_CAF1 30655 116 - 1 UUUAACGAAUCAUCGAUUAAACA-GAGAUCGUAGA--AAUUCAUUACCGGAUUUGUCAACAGCCAUUUCCGAUAAAUGAGUUAUCUGCGAAUGGCGGCGAUUGCCGCUGAC-GGCUUUCU ((((((((....))).))))).(-(((((((((((--((((((((..((((..((.(....).))..))))...)))))))).)))))))..((((((....))))))...-...))))) ( -36.20) >DroSim_CAF1 14689 116 - 1 UUUAACGAAUCAUCGAUUAAACA-GAGAUCGUAGA--AAUUCAUUACCGGAUUUGUCAACAGCCAUUUCCGAUAAAUGAGUUAUCUGCGAAUGGCGGCGAUUGCCGCUGAC-GGCUUUCU ((((((((....))).))))).(-(((((((((((--((((((((..((((..((.(....).))..))))...)))))))).)))))))..((((((....))))))...-...))))) ( -36.20) >DroEre_CAF1 24704 114 - 1 AUGAACGUAUCAUCGAUUAAACA-CAGAUCGUAGA--AAUUCAUUACCGGAUUUGUCAACAAGCAUUUGCGAUAAAUGAGUUAUCUGCGAGUGC---CGAUUGCCGCCGACUGGCUUUCU .(((.((......)).)))..((-(...(((((((--((((((((..((.(..(((......)))..).))...)))))))).)))))))))).---.((..((((.....))))..)). ( -26.80) >DroYak_CAF1 19919 113 - 1 AUUAACGCACAAUCGAUUAAACA-GAGAUCGUAGA--AAUUCAUCACCGGAUUUGUCAACGAGCAUUUCCGAUAAAUGAGUUAUCUGCGAAUGG---CGAUUACCGCUGAC-GGCUUUCU .....(((.((.((((((.....-..))))(((((--(((((((...((((..(((......)))..))))....))))))).))))))).)))---))....(((....)-))...... ( -29.70) >DroPer_CAF1 17000 90 - 1 A--------------AAUAAGCA-CAAAAGGAGGAAAUAUACAUUAACAGCUCUGUCAGCGACAAUUUCCGAUA--UGUGUUGCCUG------------AUUGUUGGUGAC-AUUUCCAU .--------------........-........((((((...((((((((.....((((((((((..........--..))))).)))------------))))))))))..-)))))).. ( -20.90) >consensus AUUAACGAAUCAUCGAUUAAACA_GAGAUCGUAGA__AAUUCAUUACCGGAUUUGUCAACAACCAUUUCCGAUAAAUGAGUUAUCUGCGAAUGG___CGAUUGCCGCUGAC_GGCUUUCU ............................(((((((...(((((((..((((..((........))..))))...)))))))..)))))))...............(((....)))..... (-13.84 = -15.07 + 1.23)

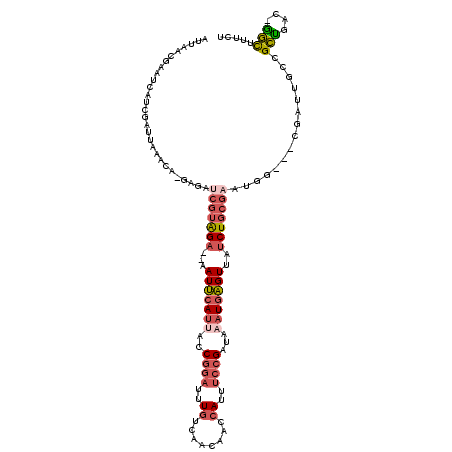
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:35:06 2006