
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,497,549 – 14,497,751 |
| Length | 202 |
| Max. P | 0.999804 |

| Location | 14,497,549 – 14,497,657 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 96.85 |
| Mean single sequence MFE | -29.86 |
| Consensus MFE | -25.78 |
| Energy contribution | -26.02 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.86 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14497549 108 + 22407834 UGUUAUAAAUUACACCCAGAGCCAUUACAAUUUUUCGUAUUCUCGGCUCAAGUUGCCGGUCUCUGAGGGCGGCACACCCAUUUGGGGCGUCAAUCAUGCGUAAAGUGA .........((((.(((((((((..(((........))).....)))))..((.((((.((.....)).)))))).......))))((((.....))))))))..... ( -30.10) >DroSec_CAF1 29272 108 + 1 UGUUAUAAAUUACACCCAGAGCCAUUACAAUUUUUCGUAUUCUCGGCUCAAGUUGCCGGUCUCUGAGGGCGGCACACCCAUUUGGGGCGUCAAUCAUGCGUAAAGUGA .........((((.(((((((((..(((........))).....)))))..((.((((.((.....)).)))))).......))))((((.....))))))))..... ( -30.10) >DroSim_CAF1 13305 108 + 1 UGUUAUAAAUUACACCCAGAGCCAUUACAAUUUUUCGUAUUCUCGGCUCAAGUUGCCGGUCUCUGAUGGCGGCACACCCAUUUGGGGCGUCAAUCAUGCGUAAAGUGA .........((((.(((((((((..(((........))).....)))))..(((((((........))))))).........))))((((.....))))))))..... ( -29.70) >DroEre_CAF1 23283 108 + 1 UGUUAUAAAUUACACCCAAAGCCAUUACAAUUUUUCGUGUUCUCGGCUCAAGUUGCCGCACUCUUAGGGCGGCACACCCAUUUGGGGCGUCAAUCAUGCGUAAAGUGA .........((((.(((((((((...(((........)))....)))....((.(((((.((....))))))))).....))))))((((.....))))))))..... ( -28.60) >DroYak_CAF1 18506 107 + 1 UGUUAUAAAUUACACCCAAAGCCAUUACAAUUUUUCGUAUUCUCGGCUCAAGUUGCCGCUC-CUUAGGGCGGCACACCCAUUUGGGGCGUCAAUCAUGCGUAAAGUGA .........((((.(((((((((..(((........))).....)))....((.(((((((-....))))))))).....))))))((((.....))))))))..... ( -30.80) >consensus UGUUAUAAAUUACACCCAGAGCCAUUACAAUUUUUCGUAUUCUCGGCUCAAGUUGCCGGUCUCUGAGGGCGGCACACCCAUUUGGGGCGUCAAUCAUGCGUAAAGUGA .........((((.(((((((((..(((........))).....)))))..((((((..........)))))).........))))((((.....))))))))..... (-25.78 = -26.02 + 0.24)


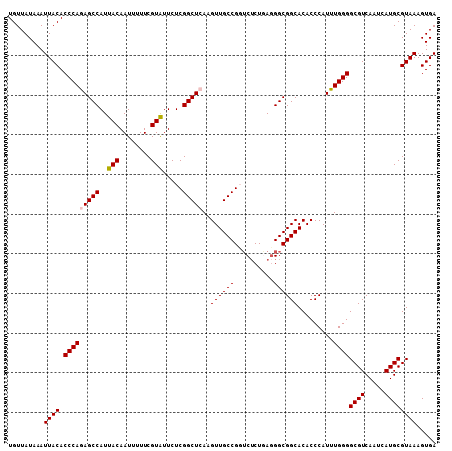
| Location | 14,497,549 – 14,497,657 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 96.85 |
| Mean single sequence MFE | -32.22 |
| Consensus MFE | -30.04 |
| Energy contribution | -30.00 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.98 |
| SVM RNA-class probability | 0.984489 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14497549 108 - 22407834 UCACUUUACGCAUGAUUGACGCCCCAAAUGGGUGUGCCGCCCUCAGAGACCGGCAACUUGAGCCGAGAAUACGAAAAAUUGUAAUGGCUCUGGGUGUAAUUUAUAACA .....((((((.......((((((.....))))))((((..(.....)..))))..((.((((((....(((((....))))).)))))).))))))))......... ( -31.40) >DroSec_CAF1 29272 108 - 1 UCACUUUACGCAUGAUUGACGCCCCAAAUGGGUGUGCCGCCCUCAGAGACCGGCAACUUGAGCCGAGAAUACGAAAAAUUGUAAUGGCUCUGGGUGUAAUUUAUAACA .....((((((.......((((((.....))))))((((..(.....)..))))..((.((((((....(((((....))))).)))))).))))))))......... ( -31.40) >DroSim_CAF1 13305 108 - 1 UCACUUUACGCAUGAUUGACGCCCCAAAUGGGUGUGCCGCCAUCAGAGACCGGCAACUUGAGCCGAGAAUACGAAAAAUUGUAAUGGCUCUGGGUGUAAUUUAUAACA .....((((((.......((((((.....)))))).......((((((.(((((.......))).....(((((....)))))..))))))))))))))......... ( -30.80) >DroEre_CAF1 23283 108 - 1 UCACUUUACGCAUGAUUGACGCCCCAAAUGGGUGUGCCGCCCUAAGAGUGCGGCAACUUGAGCCGAGAACACGAAAAAUUGUAAUGGCUUUGGGUGUAAUUUAUAACA .....((((((.......((((((.....))))))(((((.(.....).)))))..((.((((((...(((........)))..)))))).))))))))......... ( -32.50) >DroYak_CAF1 18506 107 - 1 UCACUUUACGCAUGAUUGACGCCCCAAAUGGGUGUGCCGCCCUAAG-GAGCGGCAACUUGAGCCGAGAAUACGAAAAAUUGUAAUGGCUUUGGGUGUAAUUUAUAACA .....((((((.......((((((.....))))))(((((((...)-).)))))..((.((((((....(((((....))))).)))))).))))))))......... ( -35.00) >consensus UCACUUUACGCAUGAUUGACGCCCCAAAUGGGUGUGCCGCCCUCAGAGACCGGCAACUUGAGCCGAGAAUACGAAAAAUUGUAAUGGCUCUGGGUGUAAUUUAUAACA .....((((((.......((((((.....))))))((((...........))))..((.((((((....(((((....))))).)))))).))))))))......... (-30.04 = -30.00 + -0.04)

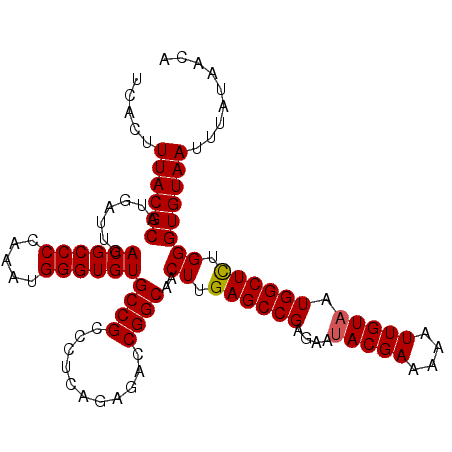
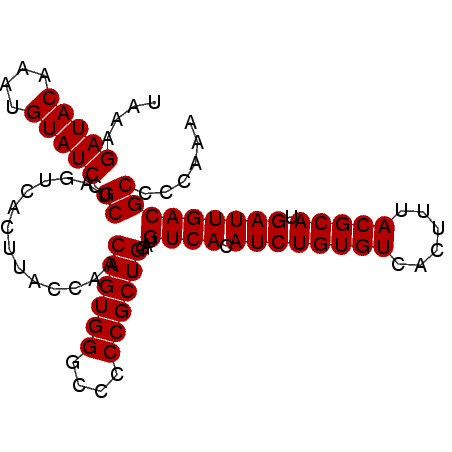
| Location | 14,497,629 – 14,497,723 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 100.00 |
| Mean single sequence MFE | -26.20 |
| Consensus MFE | -26.20 |
| Energy contribution | -26.20 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.55 |
| Structure conservation index | 1.00 |
| SVM decision value | 3.26 |
| SVM RNA-class probability | 0.998876 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14497629 94 - 22407834 UAAAGAUACAAAUGUAUCCGCUAGUCACUUACCAACAGUGGGCCCCCGCUGGAUGUCACAUCUGUGUCACUUUACGCAUGAUUGACGCCCCAAA ....(((((....))))).((..............((((((....))))))...((((.((((((((......))))).)))))))))...... ( -26.20) >DroSec_CAF1 29352 94 - 1 UAAAGAUACAAAUGUAUCCGCUAGUCACUUACCAACAGUGGGCCCCCGCUGGAUGUCACAUCUGUGUCACUUUACGCAUGAUUGACGCCCCAAA ....(((((....))))).((..............((((((....))))))...((((.((((((((......))))).)))))))))...... ( -26.20) >DroSim_CAF1 13385 94 - 1 UAAAGAUACAAAUGUAUCCGCUAGUCACUUACCAACAGUGGGCCCCCGCUGGAUGUCACAUCUGUGUCACUUUACGCAUGAUUGACGCCCCAAA ....(((((....))))).((..............((((((....))))))...((((.((((((((......))))).)))))))))...... ( -26.20) >DroEre_CAF1 23363 94 - 1 UAAAGAUACAAAUGUAUCCGCUAGUCACUUACCAACAGUGGGCCCCCGCUGGAUGUCACAUCUGUGUCACUUUACGCAUGAUUGACGCCCCAAA ....(((((....))))).((..............((((((....))))))...((((.((((((((......))))).)))))))))...... ( -26.20) >DroYak_CAF1 18585 94 - 1 UAAAGAUACAAAUGUAUCCGCUAGUCACUUACCAACAGUGGGCCCCCGCUGGAUGUCACAUCUGUGUCACUUUACGCAUGAUUGACGCCCCAAA ....(((((....))))).((..............((((((....))))))...((((.((((((((......))))).)))))))))...... ( -26.20) >consensus UAAAGAUACAAAUGUAUCCGCUAGUCACUUACCAACAGUGGGCCCCCGCUGGAUGUCACAUCUGUGUCACUUUACGCAUGAUUGACGCCCCAAA ....(((((....))))).((..............((((((....))))))...((((.((((((((......))))).)))))))))...... (-26.20 = -26.20 + -0.00)


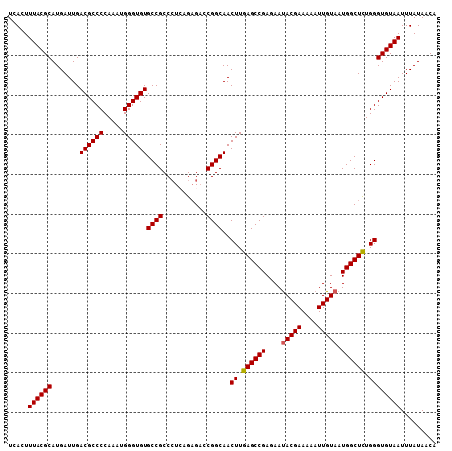
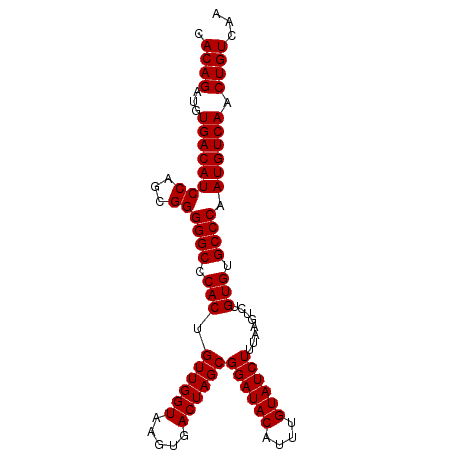
| Location | 14,497,657 – 14,497,751 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 99.57 |
| Mean single sequence MFE | -31.18 |
| Consensus MFE | -30.62 |
| Energy contribution | -30.62 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.98 |
| SVM decision value | 2.72 |
| SVM RNA-class probability | 0.996579 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14497657 94 + 22407834 CACAGAUGUGACAUCCAGCGGGGGCCCACUGUUGGUAAGUGACUAGCGGAUACAUUUGUAUCUUUAAGUCUGUGUGCCCAAUGUCAACUGUCAA .((((...((((((((...))((((.(((.((((((.....))))))((((((....))))))........))).)))).)))))).))))... ( -30.60) >DroSec_CAF1 29380 94 + 1 CACAGAUGUGACAUCCAGCGGGGGCCCACUGUUGGUAAGUGACUAGCGGAUACAUUUGUAUCUUUAAGUCUGUGUGCCCAAUGUCAACUGUCAA .((((...((((((((...))((((.(((.((((((.....))))))((((((....))))))........))).)))).)))))).))))... ( -30.60) >DroSim_CAF1 13413 94 + 1 CACAGAUGUGACAUCCAGCGGGGGCCCACUGUUGGUAAGUGACUAGCGGAUACAUUUGUAUCUUUAAGUCUGUGUGCCCAAUGUCAACUGUCAA .((((...((((((((...))((((.(((.((((((.....))))))((((((....))))))........))).)))).)))))).))))... ( -30.60) >DroEre_CAF1 23391 94 + 1 CACAGAUGUGACAUCCAGCGGGGGCCCACUGUUGGUAAGUGACUAGCGGAUACAUUUGUAUCUUUAAGUCUGUGUGCCCAAUGUCAACUGUCAA .((((...((((((((...))((((.(((.((((((.....))))))((((((....))))))........))).)))).)))))).))))... ( -30.60) >DroYak_CAF1 18613 94 + 1 CACAGAUGUGACAUCCAGCGGGGGCCCACUGUUGGUAAGUGACUAGCGGAUACAUUUGUAUCUUUAAGUCUGUGUGCCCGAUGUCAACUGUCAA .((((...((((((((((.((....)).)))..((((...((((.(.((((((....)))))).).))))....)))).))))))).))))... ( -33.50) >consensus CACAGAUGUGACAUCCAGCGGGGGCCCACUGUUGGUAAGUGACUAGCGGAUACAUUUGUAUCUUUAAGUCUGUGUGCCCAAUGUCAACUGUCAA .((((...((((((((...))((((.(((.((((((.....))))))((((((....))))))........))).)))).)))))).))))... (-30.62 = -30.62 + -0.00)


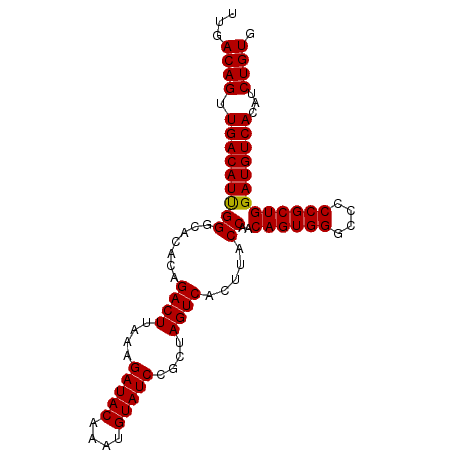
| Location | 14,497,657 – 14,497,751 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 99.57 |
| Mean single sequence MFE | -32.50 |
| Consensus MFE | -32.66 |
| Energy contribution | -32.50 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.38 |
| Structure conservation index | 1.00 |
| SVM decision value | 4.12 |
| SVM RNA-class probability | 0.999804 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14497657 94 - 22407834 UUGACAGUUGACAUUGGGCACACAGACUUAAAGAUACAAAUGUAUCCGCUAGUCACUUACCAACAGUGGGCCCCCGCUGGAUGUCACAUCUGUG ...((((.(((((((((.......((((....(((((....)))))....)))).....))..((((((....)))))))))))))...)))). ( -32.00) >DroSec_CAF1 29380 94 - 1 UUGACAGUUGACAUUGGGCACACAGACUUAAAGAUACAAAUGUAUCCGCUAGUCACUUACCAACAGUGGGCCCCCGCUGGAUGUCACAUCUGUG ...((((.(((((((((.......((((....(((((....)))))....)))).....))..((((((....)))))))))))))...)))). ( -32.00) >DroSim_CAF1 13413 94 - 1 UUGACAGUUGACAUUGGGCACACAGACUUAAAGAUACAAAUGUAUCCGCUAGUCACUUACCAACAGUGGGCCCCCGCUGGAUGUCACAUCUGUG ...((((.(((((((((.......((((....(((((....)))))....)))).....))..((((((....)))))))))))))...)))). ( -32.00) >DroEre_CAF1 23391 94 - 1 UUGACAGUUGACAUUGGGCACACAGACUUAAAGAUACAAAUGUAUCCGCUAGUCACUUACCAACAGUGGGCCCCCGCUGGAUGUCACAUCUGUG ...((((.(((((((((.......((((....(((((....)))))....)))).....))..((((((....)))))))))))))...)))). ( -32.00) >DroYak_CAF1 18613 94 - 1 UUGACAGUUGACAUCGGGCACACAGACUUAAAGAUACAAAUGUAUCCGCUAGUCACUUACCAACAGUGGGCCCCCGCUGGAUGUCACAUCUGUG ...((((.(((((((((.......((((....(((((....)))))....)))).....))..((((((....)))))))))))))...)))). ( -34.50) >consensus UUGACAGUUGACAUUGGGCACACAGACUUAAAGAUACAAAUGUAUCCGCUAGUCACUUACCAACAGUGGGCCCCCGCUGGAUGUCACAUCUGUG ...((((.(((((((((.......((((....(((((....)))))....)))).....))..((((((....)))))))))))))...)))). (-32.66 = -32.50 + -0.16)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:34:59 2006