
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,490,767 – 14,490,907 |
| Length | 140 |
| Max. P | 0.987655 |

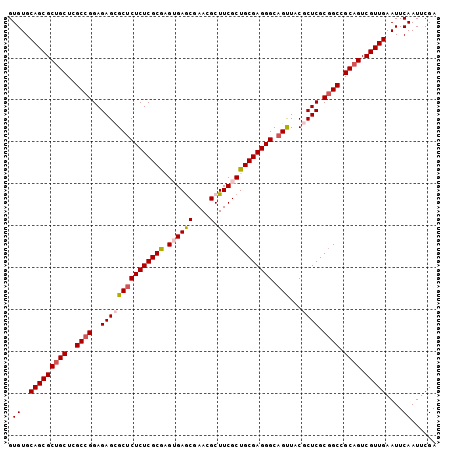
| Location | 14,490,767 – 14,490,869 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 94.31 |
| Mean single sequence MFE | -46.56 |
| Consensus MFE | -39.34 |
| Energy contribution | -40.02 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.84 |
| SVM decision value | 2.09 |
| SVM RNA-class probability | 0.987655 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14490767 102 + 22407834 GUGUGCAGCGCUGCUCGCCGGAGAGCGCUCUCUCGCGAGUGAGCGAACGCUUCGCUGCGAGGGCAGUUACGCUCGCUGCCGCAGUCGUUGAAUUCAAUUCGA (((.(((((...((((((.(((((....))))).))))))((((((((.(((((...)))))...))).)))))))))))))....((((....)))).... ( -44.90) >DroSec_CAF1 22492 102 + 1 GUGUGCAGCGCUGCUCGCCGGAGAGCGCUCUCUCGCAAGUGAGCGAACGCUUCGCUGCGAGGGCAGCUACGCUCGCGGCCGCAGUCGUUGAAUUCAAUUCGA .((..(((((((((..((((..(((((((((((((((.((((((....)).)))))))))))).)))...)))).)))).)))).)))))....))...... ( -50.10) >DroSim_CAF1 6536 102 + 1 GUGUGCAGCGCUGCUCGCCGGAGAGCGCUCUCUCGCAAGUGAGCGAACGCUUCGCUGCGAGGGCAGUUACGCUCGCGGCCGCAGUCGUUGAAUUCAAUUCGA .((..(((((((((..((((..(((((((((((((((.((((((....)).)))))))))))).))...))))).)))).)))).)))))....))...... ( -49.10) >DroEre_CAF1 16412 102 + 1 GUGUGCAGCGCUGCUCGCCGGAGAGAGCUCUCUCGCGUGAGAACGAACGCUUCGCUGCGAGGGCAGCUACACUCGCGGCCGCUGUCGUUGAAUUCAAUUCGA .((..((((((.((..(((((((.(((((((((((((((((.........))))).))))))).)))).).))).)))).)).).)))))....))...... ( -40.70) >DroYak_CAF1 11651 102 + 1 GUGUGCAGCGCUGCUCGCCGGAGAGAGCGCUCUCGCGUGAGAACGAACGCUUCGCUGCGAGGGCAGUUACACUCGCGGCGGCAGUCGUUGAAUUCAAUUCGA .((..(((((((((.((((((((.(((((((((((((((((.........))))).)))))))).))).).))).))))))))).)))))....))...... ( -48.00) >consensus GUGUGCAGCGCUGCUCGCCGGAGAGCGCUCUCUCGCGAGUGAGCGAACGCUUCGCUGCGAGGGCAGUUACGCUCGCGGCCGCAGUCGUUGAAUUCAAUUCGA .((..(((((((((..((((..(((((((((((((((.((((((....).))))))))))))).)))...)))).)))).)))).)))))....))...... (-39.34 = -40.02 + 0.68)


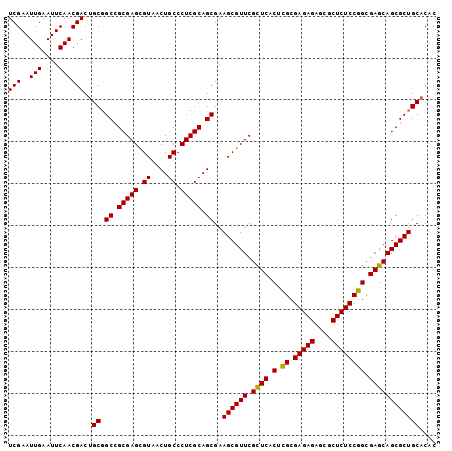
| Location | 14,490,767 – 14,490,869 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 94.31 |
| Mean single sequence MFE | -39.90 |
| Consensus MFE | -37.56 |
| Energy contribution | -37.08 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.903444 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14490767 102 - 22407834 UCGAAUUGAAUUCAACGACUGCGGCAGCGAGCGUAACUGCCCUCGCAGCGAAGCGUUCGCUCACUCGCGAGAGAGCGCUCUCCGGCGAGCAGCGCUGCACAC (((..(((....)))))).(((((((((((((((..((((....))))....)))))))))..((((((((((....)))))..)))))....))))))... ( -41.40) >DroSec_CAF1 22492 102 - 1 UCGAAUUGAAUUCAACGACUGCGGCCGCGAGCGUAGCUGCCCUCGCAGCGAAGCGUUCGCUCACUUGCGAGAGAGCGCUCUCCGGCGAGCAGCGCUGCACAC (((..(((....)))))).((((((.((((((((.(((((....)))))...))))))((((.((.(.(((((....)))))))).)))).))))))))... ( -43.40) >DroSim_CAF1 6536 102 - 1 UCGAAUUGAAUUCAACGACUGCGGCCGCGAGCGUAACUGCCCUCGCAGCGAAGCGUUCGCUCACUUGCGAGAGAGCGCUCUCCGGCGAGCAGCGCUGCACAC ............((.((.((((.((((((((.((....)).))))).(.(((((((((.(((......))).))))))).))))))..)))))).))..... ( -38.60) >DroEre_CAF1 16412 102 - 1 UCGAAUUGAAUUCAACGACAGCGGCCGCGAGUGUAGCUGCCCUCGCAGCGAAGCGUUCGUUCUCACGCGAGAGAGCUCUCUCCGGCGAGCAGCGCUGCACAC (((..(((....))))))..(((((.((((((((.(((((....)))))...))))))((((.(.((.(((((....)))))))).)))).))))))).... ( -38.50) >DroYak_CAF1 11651 102 - 1 UCGAAUUGAAUUCAACGACUGCCGCCGCGAGUGUAACUGCCCUCGCAGCGAAGCGUUCGUUCUCACGCGAGAGCGCUCUCUCCGGCGAGCAGCGCUGCACAC (((..(((....)))))).((((((.(((((.((....)).))))).))).((((((.((((((....))))))((((.(....).)))))))))))))... ( -37.60) >consensus UCGAAUUGAAUUCAACGACUGCGGCCGCGAGCGUAACUGCCCUCGCAGCGAAGCGUUCGCUCACUCGCGAGAGAGCGCUCUCCGGCGAGCAGCGCUGCACAC (((..(((....))))))..((.((.(((((.((....)).))))).))..((((((.((((.(.((.(((((....)))))))).)))))))))))).... (-37.56 = -37.08 + -0.48)

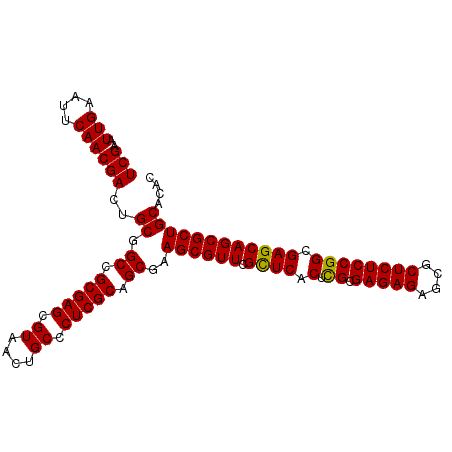
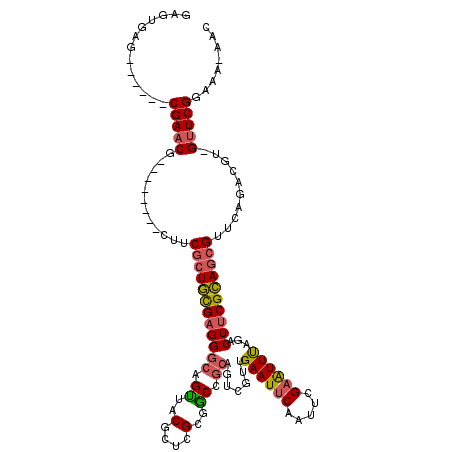
| Location | 14,490,803 – 14,490,907 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.67 |
| Mean single sequence MFE | -37.68 |
| Consensus MFE | -26.18 |
| Energy contribution | -26.10 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.551555 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14490803 104 + 22407834 GAGUGAG------CGAACG--------CUUCGCUGCGAGGGCAGUUACGCUCGCUGCCGCAGUCGUUGAAUUCAAUUCGAAUUUUGACUUCGCAGCGUUCAGACGU-GUUCGGAAA-AAC .......------((((((--------(..((((((((.((((((.......))))))..(((((..((((((.....))))))))))))))))))).......))-)))))....-... ( -39.20) >DroSec_CAF1 22528 104 + 1 AAGUGAG------CGAACG--------CUUCGCUGCGAGGGCAGCUACGCUCGCGGCCGCAGUCGUUGAAUUCAAUUCGAAUUUUGACUUCGCAGCGUUCAGACGU-GUUCGGAAA-AAC .......------((((((--------(.(((((((....))))).(((((.((((....(((((..((((((.....))))))))))))))))))))...)).))-)))))....-... ( -38.10) >DroSim_CAF1 6572 104 + 1 AAGUGAG------CGAACG--------CUUCGCUGCGAGGGCAGUUACGCUCGCGGCCGCAGUCGUUGAAUUCAAUUCGAAUUUUGACUUCGCAGCGUUCAGACGU-GUUCGGAAA-AAC .......------((((((--------(.((((((((((.(......).))))))))((((((((..((((((.....))))))))))).....)))....)).))-)))))....-... ( -36.60) >DroEre_CAF1 16448 103 + 1 GUGAGAA------CGAACG--------CUUCGCUGCGAGGGCAGCUACACUCGCGGCCGCUGUCGUUGAAUUCAAUUCGAGUUUAGACUUCGCAGCGUUCAGACGU-GUUCGGAA--AAC .......------((((((--------(.(((((((((((.......).))))))))((((((..((((((((.....)))))))).....))))))....)).))-)))))...--... ( -40.00) >DroYak_CAF1 11687 103 + 1 GUGAGAA------CGAACG--------CUUCGCUGCGAGGGCAGUUACACUCGCGGCGGCAGUCGUUGAAUUCAAUUCGAGUUUAGACUUCGCAGCGUUCAGACGU-GUUCGGAA--AAC .......------((((((--------(..((((((((((....(((.(((((((((....))))).((((...)))))))).))).)))))))))).......))-)))))...--... ( -38.10) >DroPer_CAF1 5708 120 + 1 GAGUGAGCGUGCACGAACGUGUCCGUUCUUCACUUUGCGGCUAGGUACGUUCGCACCGUCACACGGAGAAUACAGUUCGAGUUCAGCCUUCGAAUCGUUCAGACGUUGCUCGGAAAAAAG ..((((((((((..(((((....)))))....((........)))))))))))).(((.((.(((..((((...(((((((.......))))))).))))...)))))..)))....... ( -34.10) >consensus GAGUGAG______CGAACG________CUUCGCUGCGAGGGCAGUUACGCUCGCGGCCGCAGUCGUUGAAUUCAAUUCGAAUUUAGACUUCGCAGCGUUCAGACGU_GUUCGGAAA_AAC .............(((((............((((((((((((.((..(....)..)).)).......((((((.....))))))...))))))))))..........)))))........ (-26.18 = -26.10 + -0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:34:52 2006