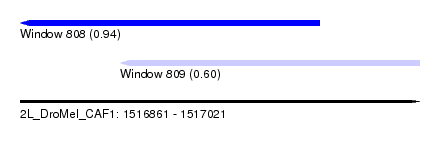
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 1,516,861 – 1,517,021 |
| Length | 160 |
| Max. P | 0.937079 |

| Location | 1,516,861 – 1,516,981 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.00 |
| Mean single sequence MFE | -33.85 |
| Consensus MFE | -27.26 |
| Energy contribution | -27.06 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.937079 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1516861 120 - 22407834 AAAUGAUUAGGCGGAAUGAAACCAACAAAUCCGCCGCCAAAAGGCCAUAUUUUCACACCAAGCCAAUUAUCCUCUUUGGAUGGUUCCUCUAUGCUGGGGAUGUGAUUUCGUAUUUUGCAU ..((((...((((((..............))))))(((....))).......(((((........(((((((.....)))))))(((((......))))))))))..))))......... ( -31.84) >DroSec_CAF1 134386 120 - 1 AAAUGAUUAAGCGGAAUAACACCAACAAAUCCGCUGCCGAGAGGCCAUAUUUUCACACCAAGCCAAUUAUCCUCUUUGGAUGGUUCCUCUUUGCUGGGGAUGUGAUUUCGUAUUUUGCAU ..((((...((((((..............))))))(((....)))((((((..((((..(((..((((((((.....))))))))...))))).))..))))))...))))......... ( -32.24) >DroSim_CAF1 137000 120 - 1 AAAUGAUUAAGCGGAAUGAAACCAACAAAUCCGCUGCCGAGAGGCCAUAUUUUCACACCAAGCCAAUUAUCCUCUUCGGAUGGUUCCUCUUUGCUGGGGAUGUGAUUUCGUAUUUUGCAU ..........(((((((((((.((....((((.(.((.((((((....................((((((((.....)))))))))))))).)).).)))).)).))))))..))))).. ( -32.05) >DroEre_CAF1 140331 120 - 1 AAAUGAUUAGGCUGAAUGAAACCAGAAAAUCCGCUGCCGAAAGGCCAUAUUUUCACACCAAGCCAAUUAUCCUUUUGGGUUGGUUCCUCCUUGCCGGGGGUGUGACUUCGUAUCUUAAAU ......(((((....(((((...............(((....))).......(((((((((.((((........)))).))))).(((((.....))))).)))).)))))..))))).. ( -32.70) >DroYak_CAF1 137774 120 - 1 AAAUGAUUAAGCUGAAUGAAACCAACAGAUCCGCGGCCGAAAGGCCACAUUUUUACACCGAGCCAAUUAUCCUUUUUGGAUGGCUCCUCUUAGUCGGGGGUGUAACUUCGUGUUUCAAAU ................(((((((.........(.((((....)))).)....(((((((((((((....(((.....)))))))))(((......))))))))))....).))))))... ( -40.40) >consensus AAAUGAUUAAGCGGAAUGAAACCAACAAAUCCGCUGCCGAAAGGCCAUAUUUUCACACCAAGCCAAUUAUCCUCUUUGGAUGGUUCCUCUUUGCUGGGGAUGUGAUUUCGUAUUUUGCAU ..((((...((((((..............))))))(((....))).......(((((....((((....(((.....)))))))(((((......))))))))))..))))......... (-27.26 = -27.06 + -0.20)

| Location | 1,516,901 – 1,517,021 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.58 |
| Mean single sequence MFE | -30.65 |
| Consensus MFE | -23.30 |
| Energy contribution | -24.18 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.603932 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1516901 120 - 22407834 GGGGACUUGACUUCCCCUGGCAUUAAAAUAUUCGUUUGGGAAAUGAUUAGGCGGAAUGAAACCAACAAAUCCGCCGCCAAAAGGCCAUAUUUUCACACCAAGCCAAUUAUCCUCUUUGGA (((((.......)))))((((....((((....)))).(((((((....((((((..............))))))(((....)))..))))))).......))))....(((.....))) ( -32.84) >DroSec_CAF1 134426 120 - 1 GGGGACUUGACUUCCCCUGCCAUUAAAAUAUUCGUUAGGGAAAUGAUUAAGCGGAAUAACACCAACAAAUCCGCUGCCGAGAGGCCAUAUUUUCACACCAAGCCAAUUAUCCUCUUUGGA ..((.((((..(((((..((.............))..)))))(((....((((((..............))))))(((....))))))..........)))))).....(((.....))) ( -28.36) >DroSim_CAF1 137040 120 - 1 GGGGACUUGACUUCCCCUGGCAUUAAAAUAUUCGUUAGGGAAAUGAUUAAGCGGAAUGAAACCAACAAAUCCGCUGCCGAGAGGCCAUAUUUUCACACCAAGCCAAUUAUCCUCUUCGGA (((((.......)))))((((.((((........))))(((((((....((((((..............))))))(((....)))..))))))).......))))....(((.....))) ( -31.74) >DroEre_CAF1 140371 120 - 1 GGGGACUUGACUUCCCCAGGCAUUAAAAUAUUCAUUUGGGAAAUGAUUAGGCUGAAUGAAACCAGAAAAUCCGCUGCCGAAAGGCCAUAUUUUCACACCAAGCCAAUUAUCCUUUUGGGU (((((.......))))).(((.........((((((..(............)..))))))....((((((.....(((....)))...)))))).......)))....((((....)))) ( -33.10) >DroYak_CAF1 137814 120 - 1 GGAGACCUUACUUCCGCGGGCAUUAAAAUAUUCAUUAACGAAAUGAUUAAGCUGAAUGAAACCAACAGAUCCGCGGCCGAAAGGCCACAUUUUUACACCGAGCCAAUUAUCCUUUUUGGA ((((.......))))(((((........((.(((((.....))))).))..(((..((....)).))).)))))((((....))))................((((.........)))). ( -27.20) >consensus GGGGACUUGACUUCCCCUGGCAUUAAAAUAUUCGUUAGGGAAAUGAUUAAGCGGAAUGAAACCAACAAAUCCGCUGCCGAAAGGCCAUAUUUUCACACCAAGCCAAUUAUCCUCUUUGGA (((((.......))))).(((.......((.(((((.....))))).))((((((..............))))))(((....)))................))).....(((.....))) (-23.30 = -24.18 + 0.88)

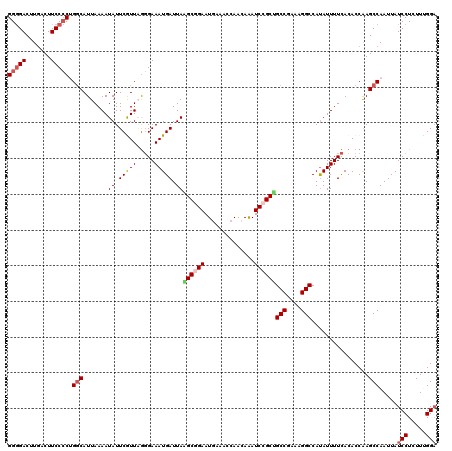
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:37:49 2006