| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,486,443 – 14,486,547 |
| Length | 104 |
| Max. P | 0.986889 |

| Location | 14,486,443 – 14,486,547 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.71 |
| Mean single sequence MFE | -40.46 |
| Consensus MFE | -30.38 |
| Energy contribution | -33.02 |
| Covariance contribution | 2.64 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.09 |
| Structure conservation index | 0.75 |
| SVM decision value | 2.06 |
| SVM RNA-class probability | 0.986889 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14486443 104 + 22407834 UUUAAUUGUCGCUCAGCUGCGCUGGACGCACGUGAUAAUUGCUCAUUUUAUGACGCAUUUAUCAAA----------UGCUCGGCCUUGGUCUCGUCCUUGGCAUUGGCAUC------GGC ......((((((......))((((((((((..((((((.(((((((...)))).))).))))))..----------)))..(((....)))..))))..)))...))))..------... ( -33.30) >DroSec_CAF1 11063 110 + 1 UUUAAUUGUCGCUCAGCUGCGCUGGACGCACGUGAUAAUUGCUCAUUUUAUGACGCAUUUAUCAAAGUGCGUUCCGAGCUCGGCCUUGGUCUCGUCCUUGGCAUUGGCAU---------- ......(((((((((((...)))((((((((.((((((.(((((((...)))).))).))))))..)))))))).))))...(((..((......))..)))...)))).---------- ( -42.00) >DroSim_CAF1 2303 110 + 1 UUUAAUUGUCGCUCAGCUGCGCUGGACGCACGUGAUAAUUGCUCAUUUUAUGACGCAUUUAUCAAAGUGCGUUCCGAGCUCGGCCUUGGUCUCGUCCUUGGCAUUGGCAU---------- ......(((((((((((...)))((((((((.((((((.(((((((...)))).))).))))))..)))))))).))))...(((..((......))..)))...)))).---------- ( -42.00) >DroEre_CAF1 12261 120 + 1 UUUAAUUGCCGCUCAGCUGCGCUGGACGCACGUGAUAAUUGCUCAUUUUAUGACGCAUUUAUCAAAGUGCGUACCGAGCUUGGCCUUGGUCUCGUCCUUGCCAUUGGCAUUGGUGUUGCC ......(((((((((((...)))((((((((.((((((.(((((((...)))).))).))))))..)))))).)))))).((((...((......))..))))..))))........... ( -43.40) >DroYak_CAF1 7201 120 + 1 UUUAAUUGUCGCUCAGCUGCGCUGGACGCACGUGAUAAUUGCUCAUUUUAUGACGCAUUUAUCAAAGUGCGUUCCGAGCUCGGCCUUCGUCUCGUCCUUGGCAUUGGAAUUGGCAUUGGC ..........((.((((((.(((((((((((.((((((.(((((((...)))).))).))))))..))))).))).))).))))....(((...(((........)))...)))..)))) ( -41.60) >consensus UUUAAUUGUCGCUCAGCUGCGCUGGACGCACGUGAUAAUUGCUCAUUUUAUGACGCAUUUAUCAAAGUGCGUUCCGAGCUCGGCCUUGGUCUCGUCCUUGGCAUUGGCAU_______G_C ......(((((((((((...)))((((((((.((((((.(((((((...)))).))).))))))..)))))))).))))...(((..((......))..)))...))))........... (-30.38 = -33.02 + 2.64)

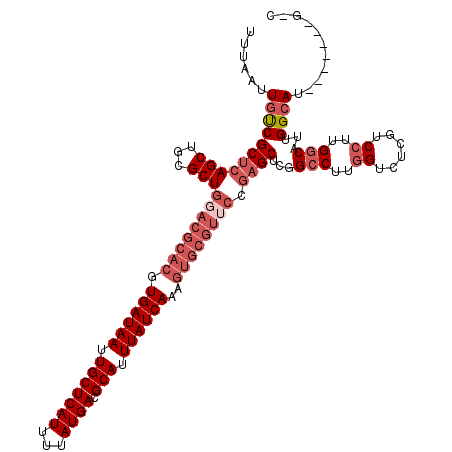
| Location | 14,486,443 – 14,486,547 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.71 |
| Mean single sequence MFE | -35.90 |
| Consensus MFE | -25.40 |
| Energy contribution | -27.60 |
| Covariance contribution | 2.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.74 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.906069 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14486443 104 - 22407834 GCC------GAUGCCAAUGCCAAGGACGAGACCAAGGCCGAGCA----------UUUGAUAAAUGCGUCAUAAAAUGAGCAAUUAUCACGUGCGUCCAGCGCAGCUGAGCGACAAUUAAA ...------(.(((((.(((...((((..(.(....).)..(((----------(.((((((.(((.((((...))))))).)))))).))))))))...)))..)).))).)....... ( -29.20) >DroSec_CAF1 11063 110 - 1 ----------AUGCCAAUGCCAAGGACGAGACCAAGGCCGAGCUCGGAACGCACUUUGAUAAAUGCGUCAUAAAAUGAGCAAUUAUCACGUGCGUCCAGCGCAGCUGAGCGACAAUUAAA ----------........(((..((......))..)))...(((((((.(((((..((((((.(((.((((...))))))).)))))).))))))))(((...))))))).......... ( -34.90) >DroSim_CAF1 2303 110 - 1 ----------AUGCCAAUGCCAAGGACGAGACCAAGGCCGAGCUCGGAACGCACUUUGAUAAAUGCGUCAUAAAAUGAGCAAUUAUCACGUGCGUCCAGCGCAGCUGAGCGACAAUUAAA ----------........(((..((......))..)))...(((((((.(((((..((((((.(((.((((...))))))).)))))).))))))))(((...))))))).......... ( -34.90) >DroEre_CAF1 12261 120 - 1 GGCAACACCAAUGCCAAUGGCAAGGACGAGACCAAGGCCAAGCUCGGUACGCACUUUGAUAAAUGCGUCAUAAAAUGAGCAAUUAUCACGUGCGUCCAGCGCAGCUGAGCGGCAAUUAAA (....).....((((..((((..((......))...)))).((((((.((((((..((((((.(((.((((...))))))).)))))).))))))))(((...)))))))))))...... ( -44.70) >DroYak_CAF1 7201 120 - 1 GCCAAUGCCAAUUCCAAUGCCAAGGACGAGACGAAGGCCGAGCUCGGAACGCACUUUGAUAAAUGCGUCAUAAAAUGAGCAAUUAUCACGUGCGUCCAGCGCAGCUGAGCGACAAUUAAA ((((.(((....(((........)))((.(.(....)))).(((.(((.(((((..((((((.(((.((((...))))))).)))))).))))))))))))))..)).)).......... ( -35.80) >consensus G_C_______AUGCCAAUGCCAAGGACGAGACCAAGGCCGAGCUCGGAACGCACUUUGAUAAAUGCGUCAUAAAAUGAGCAAUUAUCACGUGCGUCCAGCGCAGCUGAGCGACAAUUAAA ...........(((((.(((...((.(........).))..(((.((.((((((..((((((.(((.((((...))))))).)))))).))))))))))))))..)).)))......... (-25.40 = -27.60 + 2.20)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:34:45 2006