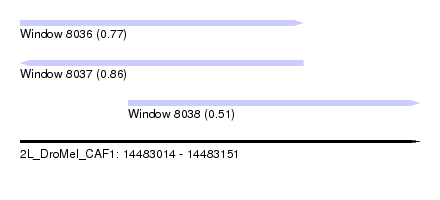
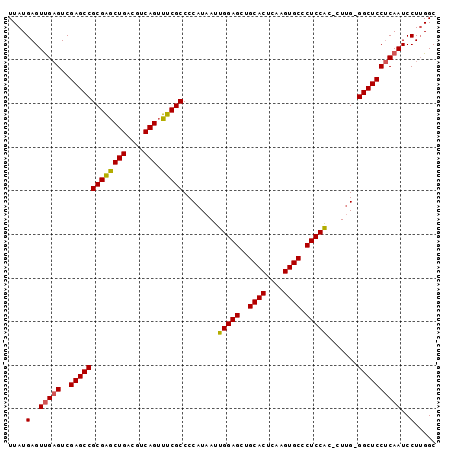
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,483,014 – 14,483,151 |
| Length | 137 |
| Max. P | 0.855254 |

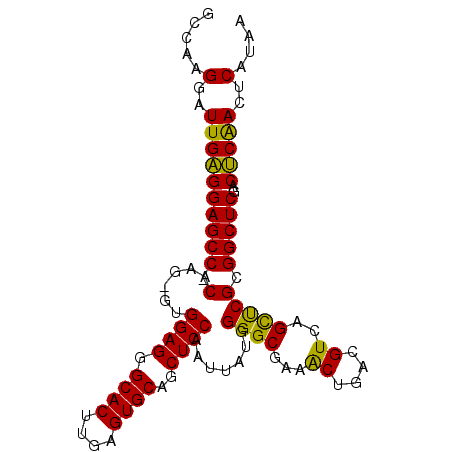
| Location | 14,483,014 – 14,483,111 |
|---|---|
| Length | 97 |
| Sequences | 3 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 92.47 |
| Mean single sequence MFE | -39.17 |
| Consensus MFE | -33.82 |
| Energy contribution | -32.50 |
| Covariance contribution | -1.32 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.770499 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14483014 97 + 22407834 GCCAAGGAUUGAGGAGCC-CAAGGGUGGAGGGCACUUGAGUGCAGCUCCAAUUAUGGGGCGAAACUGACGUCAGCUCGCGGCUCGACUCAACUCAUAA .....(..((((((((((-...((((.((.(((((....)))).((((((....))))))........).)).))))..)))))..)))))..).... ( -37.60) >DroSec_CAF1 7580 96 + 1 GCCAAGGAUUGAGGAGCC-CGAG-GUGGAGGGCACUUGAGUGCAGCUCCAAUUAUGGGGCGAAACUGACGUCAGCUCGCGGCUCGACUCAACUCAUAA .....(..((((((((((-((((-.((..(.((((....)))).((((((....))))))........)..)).)))).)))))..)))))..).... ( -41.20) >DroEre_CAF1 8990 97 + 1 GCCCAGGAUUGUGGAGCCCCAAG-GCGGAGGGCACUUGAGUGCAGCUCCAAUUAUGGGGCGAAGCUGACGUCAGUCCGCGGCUCGACGCGACUCAUAA ((...(..(((((((((((((((-..((((.((((....))))..))))..)).))))))....(((....))))))))))..)...))......... ( -38.70) >consensus GCCAAGGAUUGAGGAGCC_CAAG_GUGGAGGGCACUUGAGUGCAGCUCCAAUUAUGGGGCGAAACUGACGUCAGCUCGCGGCUCGACUCAACUCAUAA .....(..((((((((((.(......((((.((((....))))..)))).......((((...((....))..))))).)))))..)))))..).... (-33.82 = -32.50 + -1.32)

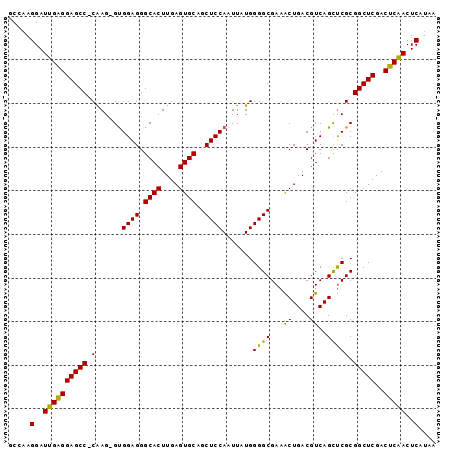
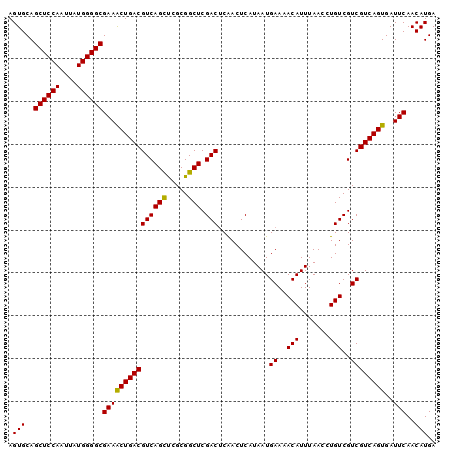
| Location | 14,483,014 – 14,483,111 |
|---|---|
| Length | 97 |
| Sequences | 3 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 92.47 |
| Mean single sequence MFE | -35.93 |
| Consensus MFE | -32.89 |
| Energy contribution | -32.90 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.855254 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14483014 97 - 22407834 UUAUGAGUUGAGUCGAGCCGCGAGCUGACGUCAGUUUCGCCCCAUAAUUGGAGCUGCACUCAAGUGCCCUCCACCCUUG-GGCUCCUCAAUCCUUGGC ....(..(((((..(((((((((((((....))).)))))..((....(((((..((((....)))).)))))....))-))))))))))..)..... ( -36.30) >DroSec_CAF1 7580 96 - 1 UUAUGAGUUGAGUCGAGCCGCGAGCUGACGUCAGUUUCGCCCCAUAAUUGGAGCUGCACUCAAGUGCCCUCCAC-CUCG-GGCUCCUCAAUCCUUGGC ....(..(((((..(((((((((((((....))).)))))........(((((..((((....)))).))))).-....-))))))))))..)..... ( -35.80) >DroEre_CAF1 8990 97 - 1 UUAUGAGUCGCGUCGAGCCGCGGACUGACGUCAGCUUCGCCCCAUAAUUGGAGCUGCACUCAAGUGCCCUCCGC-CUUGGGGCUCCACAAUCCUGGGC ....((((.((((((..(....)..))))))..)))).((((((....(((((..((((....)))).))))).-..))))))((((......)))). ( -35.70) >consensus UUAUGAGUUGAGUCGAGCCGCGAGCUGACGUCAGUUUCGCCCCAUAAUUGGAGCUGCACUCAAGUGCCCUCCAC_CUUG_GGCUCCUCAAUCCUUGGC ....(..(((((..(((((((((((((....))).)))))........(((((..((((....)))).))))).......))))))))))..)..... (-32.89 = -32.90 + 0.01)

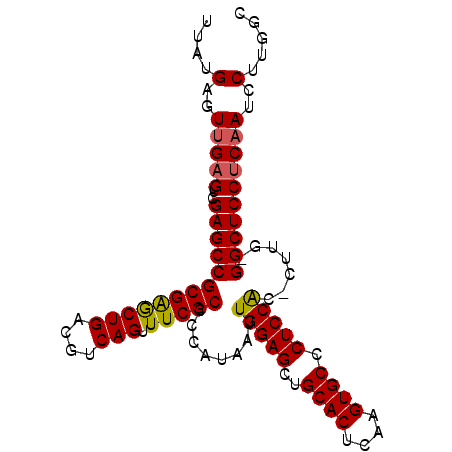
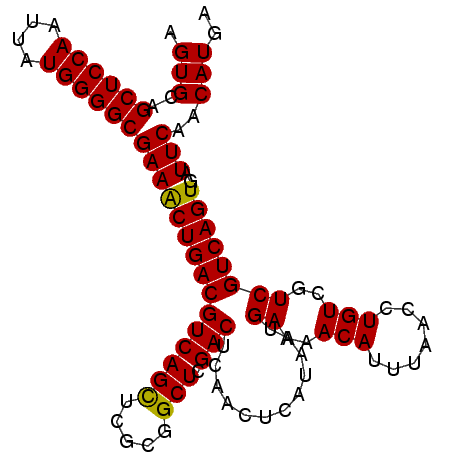
| Location | 14,483,051 – 14,483,151 |
|---|---|
| Length | 100 |
| Sequences | 3 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 96.67 |
| Mean single sequence MFE | -28.43 |
| Consensus MFE | -27.37 |
| Energy contribution | -26.93 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.96 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.507090 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14483051 100 + 22407834 AGUGCAGCUCCAAUUAUGGGGCGAAACUGACGUCAGCUCGCGGCUCGACUCAACUCAUAAUGAAAACAUUUAACCUGUCGUCGUCAGUGAUUCAACAUGA .(((..((((((....))))))(((((((((((((((.....))).)))............((..(((.......)))..))))))))..)))..))).. ( -27.00) >DroSec_CAF1 7616 100 + 1 AGUGCAGCUCCAAUUAUGGGGCGAAACUGACGUCAGCUCGCGGCUCGACUCAACUCAUAAUGAAAACAUUUAACCUGUCGUCGUCAGUGAUUCAACAUGA .(((..((((((....))))))(((((((((((((((.....))).)))............((..(((.......)))..))))))))..)))..))).. ( -27.00) >DroEre_CAF1 9027 100 + 1 AGUGCAGCUCCAAUUAUGGGGCGAAGCUGACGUCAGUCCGCGGCUCGACGCGACUCAUAAUGAAAACAUUUAACCUGUCGUCGUCAGUGAUUCAACAUGA .(((..((((((....))))))(((((((((((((((.....))).)))(((((....((((....))))......))))).))))))..)))..))).. ( -31.30) >consensus AGUGCAGCUCCAAUUAUGGGGCGAAACUGACGUCAGCUCGCGGCUCGACUCAACUCAUAAUGAAAACAUUUAACCUGUCGUCGUCAGUGAUUCAACAUGA .(((..((((((....))))))(((((((((((((((.....))).)))............((..(((.......)))..))))))))..)))..))).. (-27.37 = -26.93 + -0.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:34:42 2006