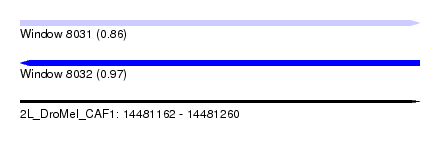
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,481,162 – 14,481,260 |
| Length | 98 |
| Max. P | 0.965724 |

| Location | 14,481,162 – 14,481,260 |
|---|---|
| Length | 98 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.99 |
| Mean single sequence MFE | -29.80 |
| Consensus MFE | -23.98 |
| Energy contribution | -23.98 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.858094 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
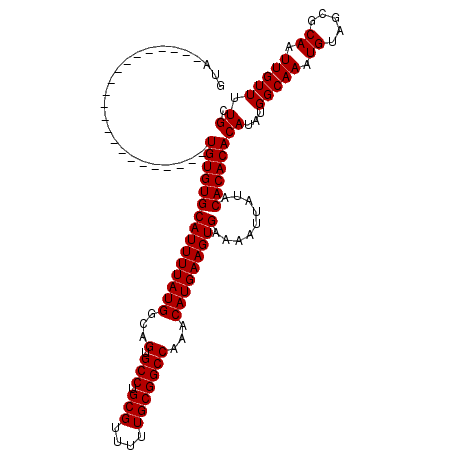
>2L_DroMel_CAF1 14481162 98 + 22407834 GUA----------------------UGUGUGCAUUUUAUGGCAGUGCCUGCGUUUUUGCGGCCAAACAUGAAGUGAAAAUUAUGCACACACAUAUGGCAAAUGUAUCGCAAUUGUUUUGC (((----------------------(((((((((((((((...(.(((.(((....)))))))...)))))))))...........)))))))))(((((.((.....)).))))).... ( -27.20) >DroSec_CAF1 5735 98 + 1 GUA----------------------UGUGUGCAUUUUAUGGCAGUGCCUGCGUUUUUGCGGCCAAACAUGAAGUGAAAAUUAUACACACACAUAUGGCAAAUGUAGCGCAAUUGUUUUGC (((----------------------(((((((((((((((...(.(((.(((....)))))))...)))))))))...........))))))))).((.......))((((.....)))) ( -27.30) >DroEre_CAF1 7244 98 + 1 GUA----------------------UGUGUGCAUUUUAUGGCAGUGCCUGCGUUUUUGCGGCCAAACAUGAAGUGAAAAUUAUACACACACAUAUGGCAAAUGUAGCGCAAUUGUUUUGC (((----------------------(((((((((((((((...(.(((.(((....)))))))...)))))))))...........))))))))).((.......))((((.....)))) ( -27.30) >DroYak_CAF1 1604 120 + 1 GUAUGUGCCGUACUAUGGGUACGUAUGUGUGCAUUUUAUGGCAGUGCCUGCGUUUUUGCGGCCAAACAUGAAGUGAAAAUUAUACACACACAUAUGGCAAAUGUAGCGCAAUUGUUUUGC .(((.((((((((.....))))((((((((((((((((((...(.(((.(((....)))))))...)))))))))...........))))))))))))).)))....((((.....)))) ( -37.40) >consensus GUA______________________UGUGUGCAUUUUAUGGCAGUGCCUGCGUUUUUGCGGCCAAACAUGAAGUGAAAAUUAUACACACACAUAUGGCAAAUGUAGCGCAAUUGUUUUGC .........................(((((((((((((((...(.(((.(((....)))))))...))))))))).........))))))((...(((((.((.....)).))))).)). (-23.98 = -23.98 + -0.00)


| Location | 14,481,162 – 14,481,260 |
|---|---|
| Length | 98 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.99 |
| Mean single sequence MFE | -25.38 |
| Consensus MFE | -21.34 |
| Energy contribution | -21.34 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.965724 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
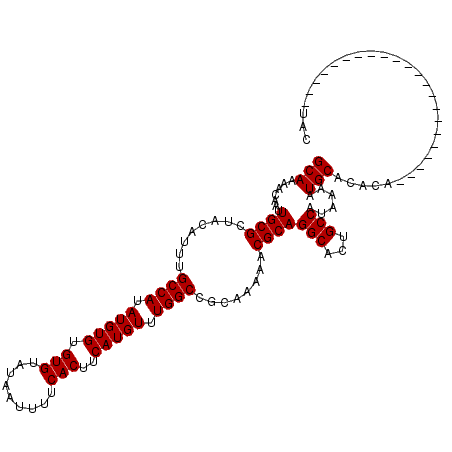
>2L_DroMel_CAF1 14481162 98 - 22407834 GCAAAACAAUUGCGAUACAUUUGCCAUAUGUGUGUGCAUAAUUUUCACUUCAUGUUUGGCCGCAAAAACGCAGGCACUGCCAUAAAAUGCACACA----------------------UAC ((((.....))))...............(((((((((((...........((....))(((((......)).)))...........)))))))))----------------------)). ( -24.20) >DroSec_CAF1 5735 98 - 1 GCAAAACAAUUGCGCUACAUUUGCCAUAUGUGUGUGUAUAAUUUUCACUUCAUGUUUGGCCGCAAAAACGCAGGCACUGCCAUAAAAUGCACACA----------------------UAC ((((.....))))((.......))....(((((((((((...........((....))(((((......)).)))...........)))))))))----------------------)). ( -22.80) >DroEre_CAF1 7244 98 - 1 GCAAAACAAUUGCGCUACAUUUGCCAUAUGUGUGUGUAUAAUUUUCACUUCAUGUUUGGCCGCAAAAACGCAGGCACUGCCAUAAAAUGCACACA----------------------UAC ((((.....))))((.......))....(((((((((((...........((....))(((((......)).)))...........)))))))))----------------------)). ( -22.80) >DroYak_CAF1 1604 120 - 1 GCAAAACAAUUGCGCUACAUUUGCCAUAUGUGUGUGUAUAAUUUUCACUUCAUGUUUGGCCGCAAAAACGCAGGCACUGCCAUAAAAUGCACACAUACGUACCCAUAGUACGGCACAUAC ((((.....))))........((((...(((((((((((...........((....))(((((......)).)))...........))))))))))).((((.....))))))))..... ( -31.70) >consensus GCAAAACAAUUGCGCUACAUUUGCCAUAUGUGUGUGUAUAAUUUUCACUUCAUGUUUGGCCGCAAAAACGCAGGCACUGCCAUAAAAUGCACACA______________________UAC (((.......((((........((((.(((((.(((.........)))..))))).))))........))))(((...)))......))).............................. (-21.34 = -21.34 + -0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:34:37 2006