| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,480,735 – 14,480,852 |
| Length | 117 |
| Max. P | 0.701103 |

| Location | 14,480,735 – 14,480,852 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.81 |
| Mean single sequence MFE | -41.54 |
| Consensus MFE | -35.58 |
| Energy contribution | -35.45 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.701103 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14480735 117 + 22407834 ---UUUGGCUCCGGCGAUUGCCAUUCCACAAUGCCCACAAAAUGGGUGCCCACUAAAAACUAAGCCACUGGCCAGCGGACGGAAAAAGUGGGUGUGGAUGUCCAUGGGGGUUGUGGAGAU ---.....(((((.(((((.(((((((((...(((((.....)))))(((((((.....((...((.((....)).))..))....)))))))))))).....)))).)))))))))).. ( -43.40) >DroSec_CAF1 5311 115 + 1 ---UUUGGCUCCGGCGAUUGCCAUUCCACAAUGCCCACAAAAUGGGUGCCCACUAAAAACUAAGCUACUGGCCAGCGGACGGAAAAAGUGGGC--UGAUGUCCAUGGGGGUUGUGGAGAU ---..((((..........))))(((((((((.((((.....((((.(((((((.........(((.......)))..........)))))))--.....)))))))).))))))))).. ( -40.81) >DroEre_CAF1 6797 120 + 1 UUGUUUGGCUCCGGCGAUUGCCAUUCCACAGUGCCCACAAAAUGGGUGCCCACUAAAAACUAAGCCACUGGCCAACGGGCGGAAAAAGUGGGUUUUGCUGUCCAUGGGGGUUGUGGAGAU ........(((((.(((((.((((...((((((((((.....)))))(((((((.........(((.(........))))......)))))))...)))))..)))).)))))))))).. ( -41.66) >DroYak_CAF1 1147 115 + 1 ---UUUGGCUCCGGCGAUUGCCAUUCCACAGUGCCCACAAAAUGGGUGCCCACUAAAAACUAAACCACUGGCCAUCGGACGGAAAAAGUGGGC--UGGUGUCCAUGGGGGAUGUGGAGAU ---.....(((((((....)))(((((...(..((((.....))))..)(((......((((..(((((..((.......))....)))))..--)))).....))))))))..)))).. ( -40.30) >consensus ___UUUGGCUCCGGCGAUUGCCAUUCCACAAUGCCCACAAAAUGGGUGCCCACUAAAAACUAAGCCACUGGCCAGCGGACGGAAAAAGUGGGC__UGAUGUCCAUGGGGGUUGUGGAGAU .....((((..........))))(((((((((.((((.....((((.(((((((.....(((......)))((.......))....))))))).......)))))))).))))))))).. (-35.58 = -35.45 + -0.13)

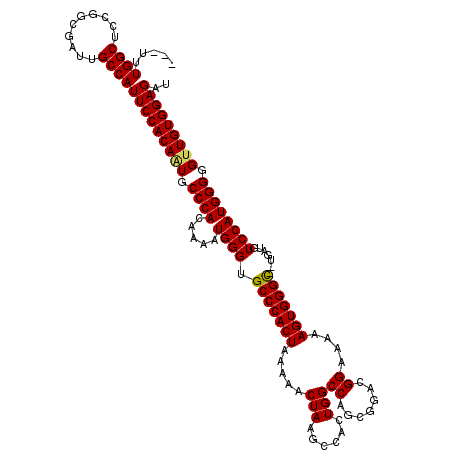

| Location | 14,480,735 – 14,480,852 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.81 |
| Mean single sequence MFE | -36.93 |
| Consensus MFE | -33.74 |
| Energy contribution | -35.12 |
| Covariance contribution | 1.37 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.631805 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14480735 117 - 22407834 AUCUCCACAACCCCCAUGGACAUCCACACCCACUUUUUCCGUCCGCUGGCCAGUGGCUUAGUUUUUAGUGGGCACCCAUUUUGUGGGCAUUGUGGAAUGGCAAUCGCCGGAGCCAAA--- ...(((((((..((((..((........((((((........(((((....)))))..........)))))).......))..))))..))))))).((((....))))........--- ( -38.53) >DroSec_CAF1 5311 115 - 1 AUCUCCACAACCCCCAUGGACAUCA--GCCCACUUUUUCCGUCCGCUGGCCAGUAGCUUAGUUUUUAGUGGGCACCCAUUUUGUGGGCAUUGUGGAAUGGCAAUCGCCGGAGCCAAA--- ...(((((((..((((..((.....--(((((((..........((((.....)))).........)))))))......))..))))..))))))).((((....))))........--- ( -39.01) >DroEre_CAF1 6797 120 - 1 AUCUCCACAACCCCCAUGGACAGCAAAACCCACUUUUUCCGCCCGUUGGCCAGUGGCUUAGUUUUUAGUGGGCACCCAUUUUGUGGGCACUGUGGAAUGGCAAUCGCCGGAGCCAAACAA ...((((((...((((..((........((((((......((((........).))).........)))))).......))..))))...)))))).((((....))))........... ( -34.12) >DroYak_CAF1 1147 115 - 1 AUCUCCACAUCCCCCAUGGACACCA--GCCCACUUUUUCCGUCCGAUGGCCAGUGGUUUAGUUUUUAGUGGGCACCCAUUUUGUGGGCACUGUGGAAUGGCAAUCGCCGGAGCCAAA--- ..((((.......((((((......--(((((((........(((........)))..........))))))).(((((...)))))..))))))...(((....))))))).....--- ( -36.07) >consensus AUCUCCACAACCCCCAUGGACAUCA__ACCCACUUUUUCCGUCCGCUGGCCAGUGGCUUAGUUUUUAGUGGGCACCCAUUUUGUGGGCACUGUGGAAUGGCAAUCGCCGGAGCCAAA___ ...(((((((..((((..((.......(((((((........(((((....)))))..........)))))))......))..))))..))))))).((((....))))........... (-33.74 = -35.12 + 1.37)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:34:35 2006