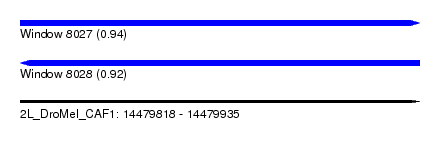
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,479,818 – 14,479,935 |
| Length | 117 |
| Max. P | 0.936024 |

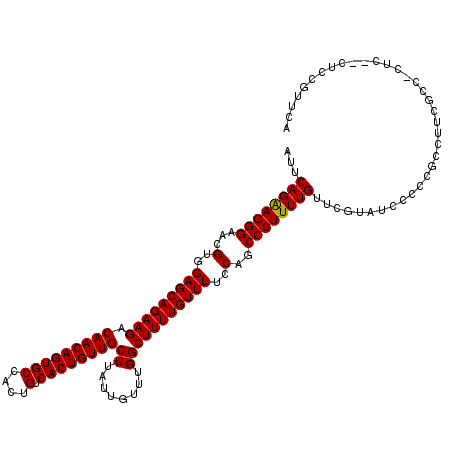
| Location | 14,479,818 – 14,479,935 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.31 |
| Mean single sequence MFE | -38.93 |
| Consensus MFE | -28.69 |
| Energy contribution | -28.63 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.15 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.936024 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14479818 117 + 22407834 UGAACAAA---GAGGGGCGAAGGCGGGGAAUACGAACAAAAGGGCUGGAAAACAAAAGCCAAACAAUAAGCAACAGUGACAGUGGCACUGUUGUCUUCUGCUCCACGUUCCCUUCUGAAU .......(---((((((((..(((((((..............((((..........)))).........(((((((((.......))))))))).)))))))...))..))))))).... ( -34.60) >DroSec_CAF1 4389 120 + 1 UGAACGGGGGGGAGUGGCGAAGGCGGGGGAUACGAACAAAAGGGCUGGAAAACAAAAGCCAAACAAUAAGCAACAGUGACAGUGGCACUGUUGUCUUCUGCUCCACGUUCCCUUCUGAAU ....(((((((((((((....((((((((.............((((..........)))).........(((((((((.......)))))))))))))))))))))..)))))))))... ( -45.00) >DroEre_CAF1 5857 105 + 1 UGAAGGGAG---------------GGGGGAUGUGAACAAAAGGGCUGGAAAACAAAAGCCAAACAAUAAGCAACAGUGACAGUGGCACUGUUGUCUUCUGCUCCACGUUCCCUCCUGAAU ....(((((---------------((((..((.((.((.(((((((..........)))).........(((((((((.......)))))))))))).)).))))..))))))))).... ( -37.20) >consensus UGAACGGAG__GAG_GGCGAAGGCGGGGGAUACGAACAAAAGGGCUGGAAAACAAAAGCCAAACAAUAAGCAACAGUGACAGUGGCACUGUUGUCUUCUGCUCCACGUUCCCUUCUGAAU ................((....))(((((((..((.((.(((((((..........)))).........(((((((((.......)))))))))))).)).))...)))))))....... (-28.69 = -28.63 + -0.05)

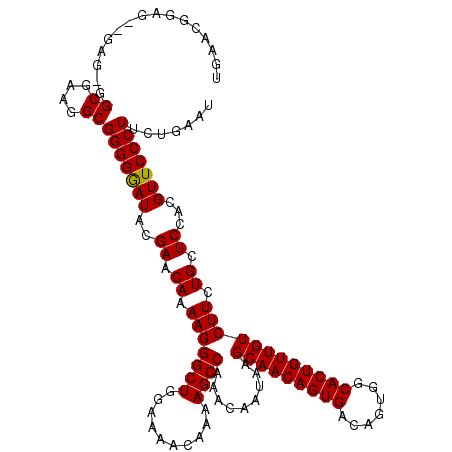

| Location | 14,479,818 – 14,479,935 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.31 |
| Mean single sequence MFE | -33.43 |
| Consensus MFE | -29.49 |
| Energy contribution | -29.27 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.923724 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14479818 117 - 22407834 AUUCAGAAGGGAACGUGGAGCAGAAGACAACAGUGCCACUGUCACUGUUGCUUAUUGUUUGGCUUUUGUUUUCCAGCCCUUUUGUUCGUAUUCCCCGCCUUCGCCCCUC---UUUGUUCA .....((((((((..((.((((((((.(((((((((....).))))))))..........((((..........))))))))))))))..))))).((....)).....---....))). ( -31.20) >DroSec_CAF1 4389 120 - 1 AUUCAGAAGGGAACGUGGAGCAGAAGACAACAGUGCCACUGUCACUGUUGCUUAUUGUUUGGCUUUUGUUUUCCAGCCCUUUUGUUCGUAUCCCCCGCCUUCGCCACUCCCCCCCGUUCA .....((((((...((((((((((((.(((((((((....).))))))))..........((((..........))))))))))))).........((....))))).....))).))). ( -31.50) >DroEre_CAF1 5857 105 - 1 AUUCAGGAGGGAACGUGGAGCAGAAGACAACAGUGCCACUGUCACUGUUGCUUAUUGUUUGGCUUUUGUUUUCCAGCCCUUUUGUUCACAUCCCCC---------------CUCCCUUCA .....((((((...((((((((((((.(((((((((....).))))))))..........((((..........))))))))))))).)))...))---------------))))..... ( -37.60) >consensus AUUCAGAAGGGAACGUGGAGCAGAAGACAACAGUGCCACUGUCACUGUUGCUUAUUGUUUGGCUUUUGUUUUCCAGCCCUUUUGUUCGUAUCCCCCGCCUUCGCC_CUC__CUCCGUUCA ...((((((((...(..(((((((((.(((((((((....).))))))))((........)))))))))))..)..)))))))).................................... (-29.49 = -29.27 + -0.22)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:34:33 2006