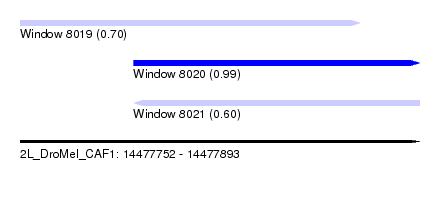
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,477,752 – 14,477,893 |
| Length | 141 |
| Max. P | 0.993282 |

| Location | 14,477,752 – 14,477,872 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.78 |
| Mean single sequence MFE | -33.00 |
| Consensus MFE | -30.47 |
| Energy contribution | -31.13 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.695226 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
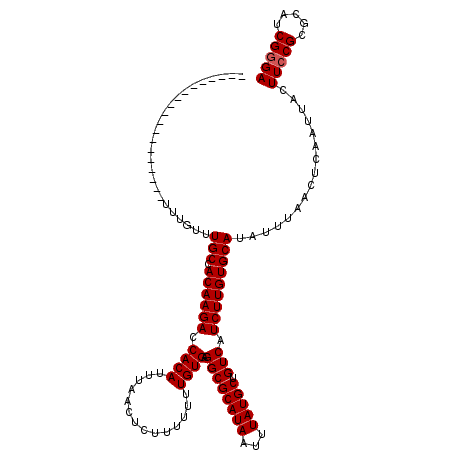
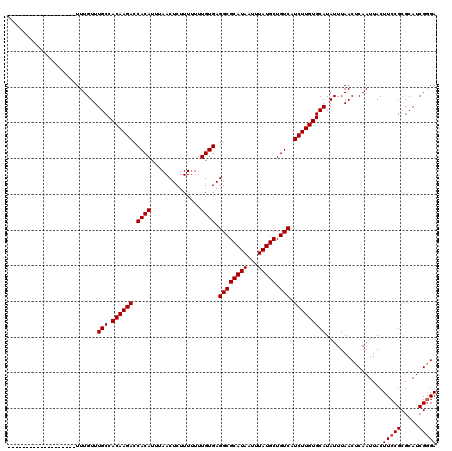
>2L_DroMel_CAF1 14477752 120 + 22407834 GCAACGUUGAUGGGGCCCACGGCCCCUAUCAACUCGGUAGUCUCGAUGCGCGGAAGUAAUUGAGUUAAAUAUGCACAAGAUGACAGCAUAAAUUAUGCGCCUCACAAAAAAAGAGUUAAA (((..((((((((((((...)))))).))))))....(((.((((((..((....)).)))))))))....)))......(((..(((((...)))))...)))................ ( -36.60) >DroSec_CAF1 2355 120 + 1 GCAACGCUGAUGGGGCCCACGGCCCCUAUCAACUCGGUAGUCCCGAUGCGCGGAAGUAAUUGAGUUAAAUAUGCACAAGAUGACAGCAUAAAUUAUGCGCCUCACAAAAAAAGAGUUAAA ........(((((((((...)))))).)))(((((((.....))((.(((((.((....))........(((((...........))))).....))))).)).........)))))... ( -32.30) >DroEre_CAF1 736 118 + 1 GCAACGUUGAUGG-GCCCACGGCCCCUAUCAACUCGGUAGUCCCGAUGCGCGGAAGUAAUUGAGUUAAAUAUGCACAAGAUGACAGCAUAAAUUAUGCGCCUCACAAA-AAAGAGUUAAA (((..((((((((-(((...)))))..))))))((((.....)))))))((....((..(((.((.......)).)))....)).(((((...)))))))(((.....-...)))..... ( -30.10) >consensus GCAACGUUGAUGGGGCCCACGGCCCCUAUCAACUCGGUAGUCCCGAUGCGCGGAAGUAAUUGAGUUAAAUAUGCACAAGAUGACAGCAUAAAUUAUGCGCCUCACAAAAAAAGAGUUAAA ..(((((((((((((((...)))))).))))))..(((.(((.((((..((....)).)))).((.......)).......))).(((((...)))))))).............)))... (-30.47 = -31.13 + 0.67)

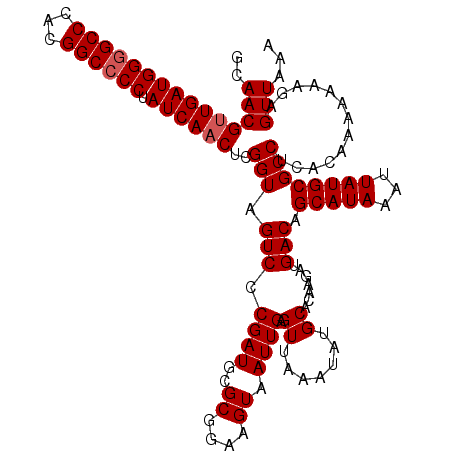
| Location | 14,477,792 – 14,477,893 |
|---|---|
| Length | 101 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.68 |
| Mean single sequence MFE | -27.03 |
| Consensus MFE | -25.43 |
| Energy contribution | -25.43 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.33 |
| Structure conservation index | 0.94 |
| SVM decision value | 2.39 |
| SVM RNA-class probability | 0.993282 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14477792 101 + 22407834 UCUCGAUGCGCGGAAGUAAUUGAGUUAAAUAUGCACAAGAUGACAGCAUAAAUUAUGCGCCUCACAAAAAAAGAGUUAAAUGUGGUCUUGUGGCAAACAAA------------------- .((((((..((....)).)))))).......((((((((((.((((((((...)))))..(((.........))).....))).))))))).)))......------------------- ( -29.30) >DroSec_CAF1 2395 101 + 1 UCCCGAUGCGCGGAAGUAAUUGAGUUAAAUAUGCACAAGAUGACAGCAUAAAUUAUGCGCCUCACAAAAAAAGAGUUAAAUGUGGUCUUGUGGCAAACAAA------------------- ...((((..((....)).)))).........((((((((((.((((((((...)))))..(((.........))).....))).))))))).)))......------------------- ( -25.50) >DroEre_CAF1 775 119 + 1 UCCCGAUGCGCGGAAGUAAUUGAGUUAAAUAUGCACAAGAUGACAGCAUAAAUUAUGCGCCUCACAAA-AAAGAGUUAAAUGUGGUCUUGUGGCAAACAAAAAAACAAAAAAAAACAGAA ...((((..((....)).)))).........((((((((((.((((((((...)))))..(((.....-...))).....))).))))))).)))......................... ( -26.30) >consensus UCCCGAUGCGCGGAAGUAAUUGAGUUAAAUAUGCACAAGAUGACAGCAUAAAUUAUGCGCCUCACAAAAAAAGAGUUAAAUGUGGUCUUGUGGCAAACAAA___________________ ...((((..((....)).)))).........((((((((((.((((((((...)))))..(((.........))).....))).))))))).)))......................... (-25.43 = -25.43 + 0.00)

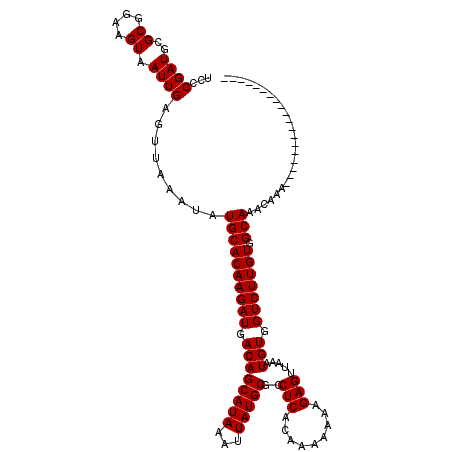

| Location | 14,477,792 – 14,477,893 |
|---|---|
| Length | 101 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.68 |
| Mean single sequence MFE | -20.18 |
| Consensus MFE | -18.81 |
| Energy contribution | -19.14 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.596861 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14477792 101 - 22407834 -------------------UUUGUUUGCCACAAGACCACAUUUAACUCUUUUUUUGUGAGGCGCAUAAUUUAUGCUGUCAUCUUGUGCAUAUUUAACUCAAUUACUUCCGCGCAUCGAGA -------------------......(((.((((((.((((..............)))).((((((((...))))).))).))))))))).......(((.................))). ( -18.27) >DroSec_CAF1 2395 101 - 1 -------------------UUUGUUUGCCACAAGACCACAUUUAACUCUUUUUUUGUGAGGCGCAUAAUUUAUGCUGUCAUCUUGUGCAUAUUUAACUCAAUUACUUCCGCGCAUCGGGA -------------------......(((.((((((.((((..............)))).((((((((...))))).))).)))))))))................(((((.....))))) ( -20.74) >DroEre_CAF1 775 119 - 1 UUCUGUUUUUUUUUGUUUUUUUGUUUGCCACAAGACCACAUUUAACUCUUU-UUUGUGAGGCGCAUAAUUUAUGCUGUCAUCUUGUGCAUAUUUAACUCAAUUACUUCCGCGCAUCGGGA ............(((..((...((.(((.((((((.((((...........-..)))).((((((((...))))).))).))))))))).))..))..)))....(((((.....))))) ( -21.52) >consensus ___________________UUUGUUUGCCACAAGACCACAUUUAACUCUUUUUUUGUGAGGCGCAUAAUUUAUGCUGUCAUCUUGUGCAUAUUUAACUCAAUUACUUCCGCGCAUCGGGA .........................(((.((((((.((((..............)))).((((((((...))))).))).)))))))))................(((((.....))))) (-18.81 = -19.14 + 0.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:34:27 2006