
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,469,621 – 14,469,750 |
| Length | 129 |
| Max. P | 0.852086 |

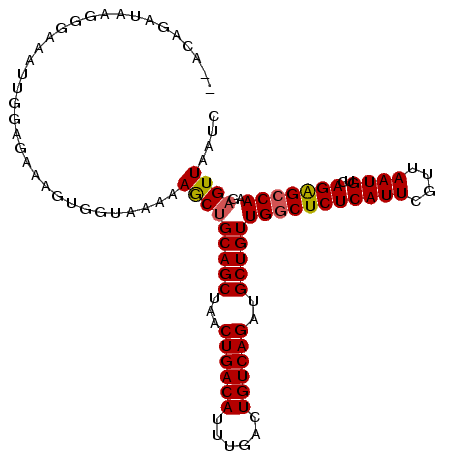
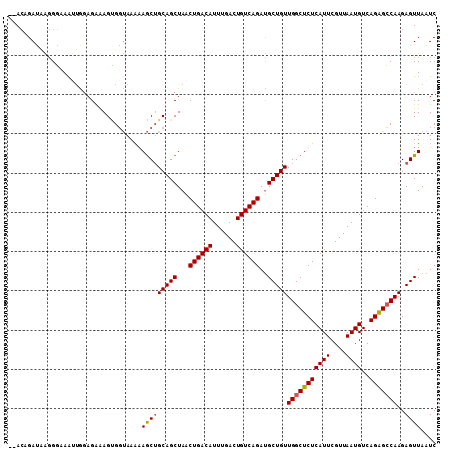
| Location | 14,469,621 – 14,469,728 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 85.19 |
| Mean single sequence MFE | -32.20 |
| Consensus MFE | -23.81 |
| Energy contribution | -23.88 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.852086 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14469621 107 + 22407834 --ACUGAUAAGGGAAAUUGGAGGGAGUGGUAAGAAGCUGCAGCUAACUGACAUUUGACUGUCAGAUGCUGUUGGCUCUCAUUCGUUAAUGUCAGGGCCAAGAGUUAAUC --.((((((.(..((((.(.((..(((.(((......))).)))..))..)))))..)))))))..(((.((((((((((((....))))..)))))))).)))..... ( -29.50) >DroSec_CAF1 56789 107 + 1 --ACAGAUAAGGGAAAUUGGAGGGAGUGGUAAGAAGCUGCAGCUAACUGACAUUUGACUGUCAGAUGCUGUUGGCUCUCAUUCGUUAAUGUCAGGGCCAAGAGUUAAUC --..............((((..(.(((........))).)..))))((((((......))))))..(((.((((((((((((....))))..)))))))).)))..... ( -28.60) >DroWil_CAF1 180234 109 + 1 AGAAAGAGCGAGUCAGAUAGAGAAAGAGCCAAAGAACAGCAGCUAACUGACAUUUGACUGUCAGAUGCUGUUGGCUCUCAUUCGUUAAUGUCAGAGGCAAGAGUUAAUC .....((..((.((.....(((..(((((((....((((((.....((((((......)))))).)))))))))))))..))).....(((.....))).)).))..)) ( -31.30) >DroAna_CAF1 47477 107 + 1 --ACAGACUGGGGCAAGGGCAGAAAGCGGUAAAGAGCUGCAGCUAACUGACAUUUGACUGUCAGACGCUGUUGGCUCUCAUUCGUUAAUGUCAGAGCCAAGAGUUAAUC --...((((..(((...((((...(((((...(((((((((((...((((((......))))))..)))).)))))))...)))))..))))...)))...)))).... ( -39.40) >consensus __ACAGAUAAGGGAAAUUGGAGAAAGUGGUAAAAAGCUGCAGCUAACUGACAUUUGACUGUCAGAUGCUGUUGGCUCUCAUUCGUUAAUGUCAGAGCCAAGAGUUAAUC ..................................(((((((((...((((((......))))))..)))))(((((((((((....))))..)))))))..)))).... (-23.81 = -23.88 + 0.06)

| Location | 14,469,621 – 14,469,728 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 85.19 |
| Mean single sequence MFE | -25.00 |
| Consensus MFE | -21.30 |
| Energy contribution | -22.05 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.793030 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14469621 107 - 22407834 GAUUAACUCUUGGCCCUGACAUUAACGAAUGAGAGCCAACAGCAUCUGACAGUCAAAUGUCAGUUAGCUGCAGCUUCUUACCACUCCCUCCAAUUUCCCUUAUCAGU-- (((......(((((.((..((((....)))))).)))))((((..((((((......))))))...))))...............................)))...-- ( -21.30) >DroSec_CAF1 56789 107 - 1 GAUUAACUCUUGGCCCUGACAUUAACGAAUGAGAGCCAACAGCAUCUGACAGUCAAAUGUCAGUUAGCUGCAGCUUCUUACCACUCCCUCCAAUUUCCCUUAUCUGU-- .........(((((.((..((((....)))))).)))))((((..((((((......))))))...)))).....................................-- ( -20.90) >DroWil_CAF1 180234 109 - 1 GAUUAACUCUUGCCUCUGACAUUAACGAAUGAGAGCCAACAGCAUCUGACAGUCAAAUGUCAGUUAGCUGCUGUUCUUUGGCUCUUUCUCUAUCUGACUCGCUCUUUCU ..........................((..((((((((((((((.((((((......)))))).....))))))....))))))))..))................... ( -28.70) >DroAna_CAF1 47477 107 - 1 GAUUAACUCUUGGCUCUGACAUUAACGAAUGAGAGCCAACAGCGUCUGACAGUCAAAUGUCAGUUAGCUGCAGCUCUUUACCGCUUUCUGCCCUUGCCCCAGUCUGU-- .....(((...(((...(.((..((((...((((((...((((..((((((......))))))...))))..))))))...)).))..)).)...)))..)))....-- ( -29.10) >consensus GAUUAACUCUUGGCCCUGACAUUAACGAAUGAGAGCCAACAGCAUCUGACAGUCAAAUGUCAGUUAGCUGCAGCUCCUUACCACUCCCUCCAAUUGCCCCUAUCUGU__ .........((((((((..((((....))))))))))))((((..((((((......))))))...))))....................................... (-21.30 = -22.05 + 0.75)

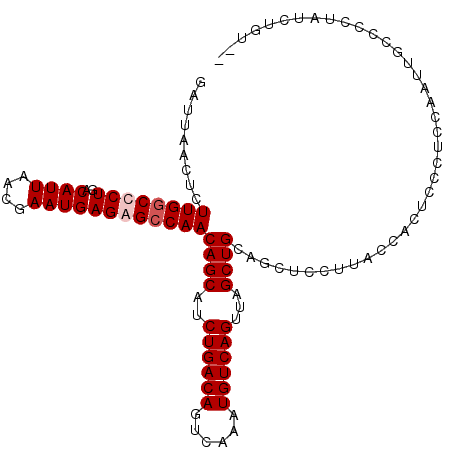
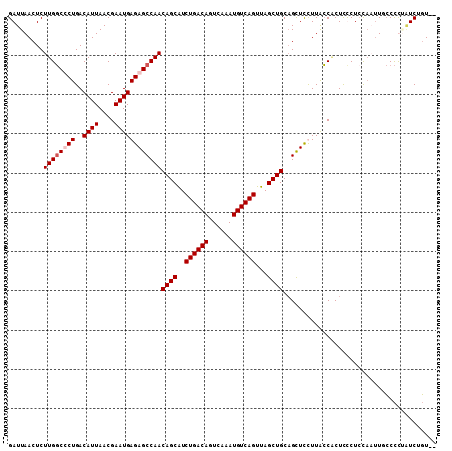
| Location | 14,469,657 – 14,469,750 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 94.43 |
| Mean single sequence MFE | -27.78 |
| Consensus MFE | -23.17 |
| Energy contribution | -23.17 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.827181 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14469657 93 + 22407834 GCAGCUAACUGACAUU-UGACUGUCAGAUGCUGUUGGCUCUCAUUCGUUAAUGUCAGGGCCAAGAGUUAAUCUCGCGUGCAUUUCGAAAGAGCC (((((...((((((..-....))))))..))))).((((((..((((..(((((..(.((...(((.....))))).)))))).)))))))))) ( -31.50) >DroPse_CAF1 76518 93 + 1 GCAGCUAACUGACAUU-UGACUGUCAGAUGCUGUUGGCUCUCAUUCGUUAAUGUCAGAGGCAAGAGUUAAUCUCGCGUGCAUUUCAAAACAACA (((((...((((((..-....))))))..(((.(((.((((((((....))))..)))).))).))).......)).))).............. ( -24.90) >DroSec_CAF1 56825 93 + 1 GCAGCUAACUGACAUU-UGACUGUCAGAUGCUGUUGGCUCUCAUUCGUUAAUGUCAGGGCCAAGAGUUAAUCUCGCGUGCAUUUCGAAAGAGCC (((((...((((((..-....))))))..))))).((((((..((((..(((((..(.((...(((.....))))).)))))).)))))))))) ( -31.50) >DroWil_CAF1 180272 93 + 1 GCAGCUAACUGACAUU-UGACUGUCAGAUGCUGUUGGCUCUCAUUCGUUAAUGUCAGAGGCAAGAGUUAAUCUCGCGUGCAUUUCACAACAGUC ........((((((..-....))))))..(((((((.((((((((....))))..))))(((.(((.....)))...)))......))))))). ( -26.00) >DroAna_CAF1 47513 93 + 1 GCAGCUAACUGACAUU-UGACUGUCAGACGCUGUUGGCUCUCAUUCGUUAAUGUCAGAGCCAAGAGUUAAUCUCGCGUGCAUUUCAAAACAACG (((((...((((((..-....))))))..(((.((((((((((((....))))..)))))))).))).......)).))).............. ( -29.60) >DroPer_CAF1 62361 94 + 1 GCAGCUAACUGACAUUUUGACUGUCAGAUGCUGUUGGCUCUCAUUCGUUAAUGUCAGAAGCAAGAGUUAAUCUCGCGUGCAUUUCAAAACAACA (((((...((((((((.(((.((..(((.((.....))))))).)))..))))))))......(((.....))))).))).............. ( -23.20) >consensus GCAGCUAACUGACAUU_UGACUGUCAGAUGCUGUUGGCUCUCAUUCGUUAAUGUCAGAGCCAAGAGUUAAUCUCGCGUGCAUUUCAAAACAACC (((((...((((((((.(((..(((((......)))))..)))......))))))))......(((.....))))).))).............. (-23.17 = -23.17 + 0.00)

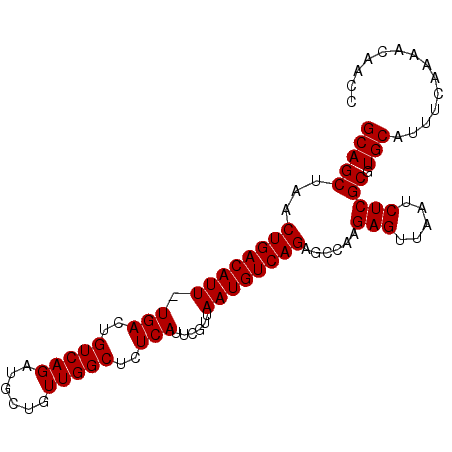
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:34:24 2006