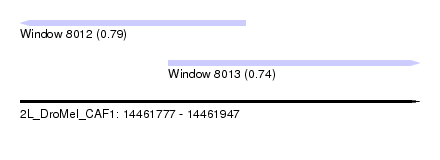
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,461,777 – 14,461,947 |
| Length | 170 |
| Max. P | 0.794668 |

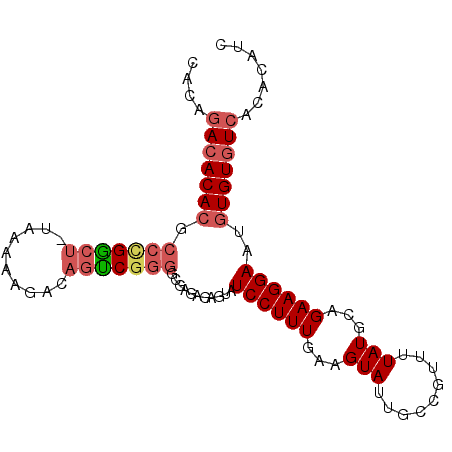
| Location | 14,461,777 – 14,461,873 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 85.53 |
| Mean single sequence MFE | -28.48 |
| Consensus MFE | -21.12 |
| Energy contribution | -22.24 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.794668 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14461777 96 - 22407834 CACAGACACACGCCCGGCUUUAAAAAGACAGUCGGGGCGAGAGAGUAUCCUUUGAAGUAUUGCCGUUUUAUGCAGAAGGAAUGUGUGUCACACAUC ....(((((((((((((((..........)))).)))).........((((((...((((.........)))).))))))..)))))))....... ( -29.80) >DroSec_CAF1 51895 95 - 1 CACAGACACACGCCCGGCU-UAAAAAGACAGUCGGGGCGAGAGAGUAUCCUUUGAAGUAUUGCCGUUUUAUGCAGAAGGAAUGUGUGUCACACAUC ....(((((((((((((((-.........)))).)))).........((((((...((((.........)))).))))))..)))))))....... ( -29.90) >DroEre_CAF1 55837 95 - 1 CCCAGACACACGCCUGGCU-UAAAAAGACAGUCGGGCCGAGAGAGUAUCCUUUGAAGUAGUGCCGUUUUAUGCAGAAGGAAUGUGUGUCACACAUC ....(((((((((((((((-.........))))))))..........((((((...((((.......))))...))))))..)))))))....... ( -31.20) >DroAna_CAF1 43540 93 - 1 CACAGACACACGCCUGGCU-GAGAGGGACAGCCGGGUAGAGAGAG--UCCUUUGAAGUAGUGCCGUAUUAUGCAGAAGGAAUGUGUGUCACACAUC ....(((((((((((((((-(.......)))))))))........--((((((...((((((...))))))...))))))..)))))))....... ( -36.90) >DroPer_CAF1 56492 88 - 1 AAACAAAACA-----GAA--AAAAAAGACAGUCGGUUGGCCAGA-UAUCCUUUGAAGUAUUGCCGUAUUAUGCAGAAGGAAUGUGUGUCACACAUC ..........-----...--......(((((((((....)).))-).((((((...((((.........)))).)))))).....))))....... ( -14.60) >consensus CACAGACACACGCCCGGCU_UAAAAAGACAGUCGGGGCGAGAGAGUAUCCUUUGAAGUAUUGCCGUUUUAUGCAGAAGGAAUGUGUGUCACACAUC ....(((((((.(((((((..........)))))))...........((((((...(((.........)))...))))))..)))))))....... (-21.12 = -22.24 + 1.12)


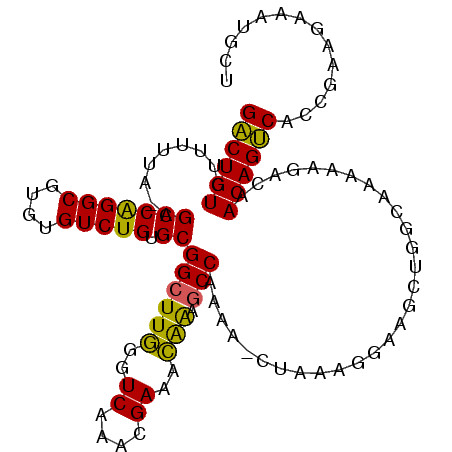
| Location | 14,461,840 – 14,461,947 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 87.07 |
| Mean single sequence MFE | -31.83 |
| Consensus MFE | -24.38 |
| Energy contribution | -23.25 |
| Covariance contribution | -1.12 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.738123 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14461840 107 + 22407834 GACUGUCUUUUUAAAGCCGGGCGUGUGUCUGUGCGGCUUGGGUCAAACGAAACAAAGCCCAAA-CUAAAGGAAGCUGGCUAAAAGACAACAGUCACCGAAGAAAUGCU ((((((..((((..((((((((.(((.((.((..(((....)))..)))).)))..))))...-....((....))))))..))))..)))))).............. ( -30.70) >DroSec_CAF1 51958 106 + 1 GACUGUCUUUUUA-AGCCGGGCGUGUGUCUGUGCGGCUUGGGUCAAACGAAACAAAGCCCAAA-CUAAAGGAAGCUGGCUAAAAGACAACAGUCACCGAACAAAUGCU ((((((.(((((.-((((((((.(((.((.((..(((....)))..)))).)))..))))...-....((....)))))).)))))..)))))).............. ( -31.00) >DroEre_CAF1 55900 106 + 1 GACUGUCUUUUUA-AGCCAGGCGUGUGUCUGGGCGGCUUGGGUCAAACGAAACAAUGCCAAAA-CCAAAGGAAGCUGGCAAAAAGACAACAGUCACCGAAGAAAUGCU ((((((((((((.-.(((((.(...(.(.(((..((((((..((....))..))).)))....-))).).)..))))))))))))))...)))).............. ( -31.00) >DroAna_CAF1 43601 107 + 1 GGCUGUCCCUCUC-AGCCAGGCGUGUGUCUGUGCGGCUUUGGUCAAACGAAGGGGACCCAAAAAUGAAAGGACGCAGCCAAAAAGACAACAGCCACCGAAGAAAUGCC ((((((...(((.-.....(((.(((((((...((..(((((((...(....).)).)))))..))...))))))))))....)))..)))))).............. ( -34.60) >consensus GACUGUCUUUUUA_AGCCAGGCGUGUGUCUGUGCGGCUUGGGUCAAACGAAACAAAGCCAAAA_CUAAAGGAAGCUGGCAAAAAGACAACAGUCACCGAAGAAAUGCU ((((((.........(((((((....))))).))((((((..((....))..))).))).............................)))))).............. (-24.38 = -23.25 + -1.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:34:19 2006