
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,456,975 – 14,457,079 |
| Length | 104 |
| Max. P | 0.963199 |

| Location | 14,456,975 – 14,457,079 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 77.74 |
| Mean single sequence MFE | -29.08 |
| Consensus MFE | -18.20 |
| Energy contribution | -18.28 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.63 |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.963199 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14456975 104 + 22407834 AUUUGUUUCGACAGUAAACUUGAUGUGUUUAUGACACAUCUUGACAGUAAGUAGCUGCUGCUUCAGCUGGCUGGCUACUGUCAGUCA--------CUUGACCUUU----UUUCAUU- .........(((.........((((((((...)))))))).((((((((..(((((((((...)))).)))))..))))))))))).--------..........----.......- ( -33.40) >DroVir_CAF1 69750 94 + 1 AUUUGUUUCGACAGUUAACUUGAUGUGUUUAUGACACGUCUUGACAGUAAGUAGCUGCUGCGGA---CU--UGGCUACUGUCAGU------GCGUGUUGACCUUU------------ .......((((((((......((((((((...))))))))((((((((((((.((....))..)---))--....))))))))).------)).)))))).....------------ ( -27.10) >DroGri_CAF1 78465 100 + 1 AUUUGUUUCGACAGUAAAGUUGAUGUGUUUAUGACACGUCUUGACACUAAGUAGCCGCCGCAGA---CUACUGGCUACUGUCCGU----GUGCGUGUUGACCUUU----UU------ .......((((((((......((((((((...))))))))...((((..((((((((.......---....))))))))....))----)))).)))))).....----..------ ( -28.10) >DroWil_CAF1 166083 101 + 1 AUUUGUUUCGACAGUAAACUUGAUGUGUUUAUGACACAUCUUGACAGUAAGUAGCUGCCGCGGA-----CUUGGCUACUGUCAGUCA-------AUUUGACCUUU----UUUUUCUC .........(((((((..(..((((((((...))))))))..)....(((((.((....))..)-----))))..))))))).(((.-------....)))....----........ ( -25.80) >DroMoj_CAF1 103405 100 + 1 AUUUGUUUCGACAGUAAAGCUGAUGUGUUUAUGACACGUCUUGACAGUAAGUAGCUGCUGCGGA---CC--UGACUACUGUCUGUAGCCGAGCGUGUUGACCUUU------------ .......((((((((..((((((((((((...))))))))...((.....))))))((((((((---(.--........)))))))))...)).)))))).....------------ ( -29.20) >DroPer_CAF1 50604 107 + 1 AUUUGUUUCGACAGUAAACUUGAUGUGUUUAUGACACAUCUUGACAGUAAGUAGCAGCCGCUGCUGUUG-CCGGCUUCUGUCAGUCA--------CUUGACCUUUUUUUUUUCAUU- .........(((((....(..((((((((...))))))))..)..(((..((((((((....)))))))-)..))).))))).(((.--------...)))...............- ( -30.90) >consensus AUUUGUUUCGACAGUAAACUUGAUGUGUUUAUGACACAUCUUGACAGUAAGUAGCUGCCGCGGA___CG_CUGGCUACUGUCAGUCA_______UGUUGACCUUU____UU______ .........(((((((.....((((((((...)))))))).............((....))..............)))))))................................... (-18.20 = -18.28 + 0.08)
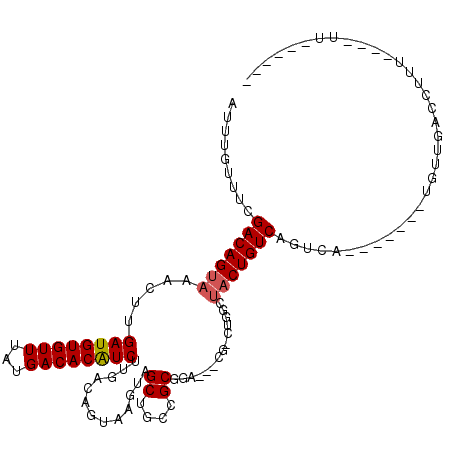

| Location | 14,456,975 – 14,457,079 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 77.74 |
| Mean single sequence MFE | -24.60 |
| Consensus MFE | -14.10 |
| Energy contribution | -14.43 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.57 |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.936888 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14456975 104 - 22407834 -AAUGAAA----AAAGGUCAAG--------UGACUGACAGUAGCCAGCCAGCUGAAGCAGCAGCUACUUACUGUCAAGAUGUGUCAUAAACACAUCAAGUUUACUGUCGAAACAAAU -.......----...((((...--------.))))((((((((..(((..((((...)))).)))..))))))))..(((((((.....)))))))..((((.......)))).... ( -30.00) >DroVir_CAF1 69750 94 - 1 ------------AAAGGUCAACACGC------ACUGACAGUAGCCA--AG---UCCGCAGCAGCUACUUACUGUCAAGACGUGUCAUAAACACAUCAAGUUAACUGUCGAAACAAAU ------------...(((.(((....------..(((((((((...--((---(........)))..))))))))).((.((((.....)))).))..))).)))............ ( -18.70) >DroGri_CAF1 78465 100 - 1 ------AA----AAAGGUCAACACGCAC----ACGGACAGUAGCCAGUAG---UCUGCGGCGGCUACUUAGUGUCAAGACGUGUCAUAAACACAUCAACUUUACUGUCGAAACAAAU ------..----................----...((((((((..(((.(---(((..((((.((....)))))).))))((((.....))))....)))))))))))......... ( -22.40) >DroWil_CAF1 166083 101 - 1 GAGAAAAA----AAAGGUCAAAU-------UGACUGACAGUAGCCAAG-----UCCGCGGCAGCUACUUACUGUCAAGAUGUGUCAUAAACACAUCAAGUUUACUGUCGAAACAAAU ........----...((((....-------.))))((((((((.(...-----.....(((((.......)))))..(((((((.....)))))))..).))))))))......... ( -28.20) >DroMoj_CAF1 103405 100 - 1 ------------AAAGGUCAACACGCUCGGCUACAGACAGUAGUCA--GG---UCCGCAGCAGCUACUUACUGUCAAGACGUGUCAUAAACACAUCAGCUUUACUGUCGAAACAAAU ------------..............(((((....((((((((...--((---(........)))..))))))))..((.((((.....)))).)).........)))))....... ( -20.40) >DroPer_CAF1 50604 107 - 1 -AAUGAAAAAAAAAAGGUCAAG--------UGACUGACAGAAGCCGG-CAACAGCAGCGGCUGCUACUUACUGUCAAGAUGUGUCAUAAACACAUCAAGUUUACUGUCGAAACAAAU -..............((((...--------.))))(((((((((.((-((..((.(((....))).))...))))..(((((((.....)))))))..)))).)))))......... ( -27.90) >consensus ______AA____AAAGGUCAACA_______UGACUGACAGUAGCCAG_AG___UCCGCAGCAGCUACUUACUGUCAAGACGUGUCAUAAACACAUCAAGUUUACUGUCGAAACAAAU ...................................((((((((.............((....)).............((.((((.....)))).))....))))))))......... (-14.10 = -14.43 + 0.33)

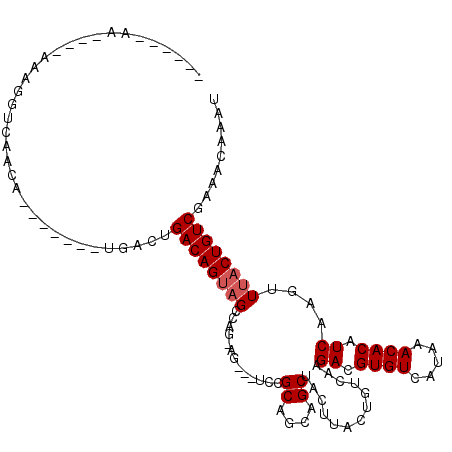

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:34:16 2006