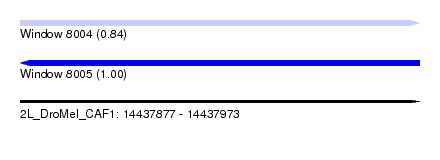
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,437,877 – 14,437,973 |
| Length | 96 |
| Max. P | 0.995152 |

| Location | 14,437,877 – 14,437,973 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 93.22 |
| Mean single sequence MFE | -25.42 |
| Consensus MFE | -21.72 |
| Energy contribution | -22.42 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.835018 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14437877 96 + 22407834 UUUUUUUUGCCCGUUUUAAUAACAGGACUCUUGCGGCGCU-UUUCGCAAUUAGCUGUCAUUCAUCAAAAUGGGACAGCUUAAUUGUGCGGUUGCUAU .........((.(((.....))).))......(((((.(.-...((((((((((((((...(((....))).)))))).)))))))).))))))... ( -24.60) >DroSec_CAF1 28131 93 + 1 UU---UUUGCCCGUUUUAAUAACAGGACUCUUGCGGCGCU-UUUCGCAAUUAGCUGUCAUUCAUCAACAUGGGACAGCUUAAUUGUGCGGUUGCUAU ..---....((.(((.....))).))......(((((.(.-...((((((((((((((...(((....))).)))))).)))))))).))))))... ( -24.50) >DroSim_CAF1 30611 93 + 1 UU---UUUGCCCGUUUUAAUAACAGGACUCUUGCGGCGCU-UUUCGCAAUUAGCUGUCAUUCAUCAAAAUGGGACAGCUUAAUUGUGCGGUUGCUAU ..---....((.(((.....))).))......(((((.(.-...((((((((((((((...(((....))).)))))).)))))))).))))))... ( -24.60) >DroEre_CAF1 30521 93 + 1 UU---UUUGCCCGCUUUAAUAACAGGACUCUCCAGGCGCU-UUUCGCAAUUAGCUGUCAUUCAUCAAAAUGGGACAGCUUAAUUGUGCGGUUGCCAU ..---...................((.....)).((((((-...((((((((((((((...(((....))).)))))).)))))))).)).)))).. ( -26.00) >DroYak_CAF1 30073 93 + 1 UU---UUUGCCCGUUUUAAUAACAGGACUCUGCAGGCGCU-UUUCGCAAUUAGCUGUCAUUCAUCAAAACGGGACAGCUUAAUUGUGCGGUUGCUAU ..---....((.(((.....))).)).....((((.(((.-....(((((((((((((..............)))))).)))))))))).))))... ( -25.94) >DroAna_CAF1 20972 94 + 1 UU---UUUGCCGGCUUUAAUAACAGGACUCUCGAGGCGCUCUUUCGCAAUUAGCUGUCAUUCAUCAAAAUGGGACAGCUUAAUUGGAAGGUGGCUAU ..---...(((((((((...............)))))...(((((.((((((((((((...(((....))).)))))).)))))))))))))))... ( -26.86) >consensus UU___UUUGCCCGUUUUAAUAACAGGACUCUUGAGGCGCU_UUUCGCAAUUAGCUGUCAUUCAUCAAAAUGGGACAGCUUAAUUGUGCGGUUGCUAU .........((.(((.....))).))........((((((....((((((((((((((...(((....))).)))))).)))))))).))).))).. (-21.72 = -22.42 + 0.70)

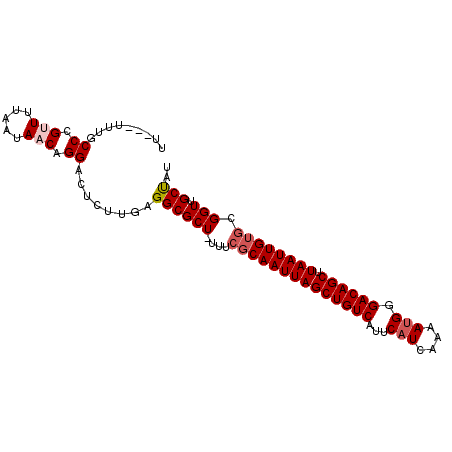
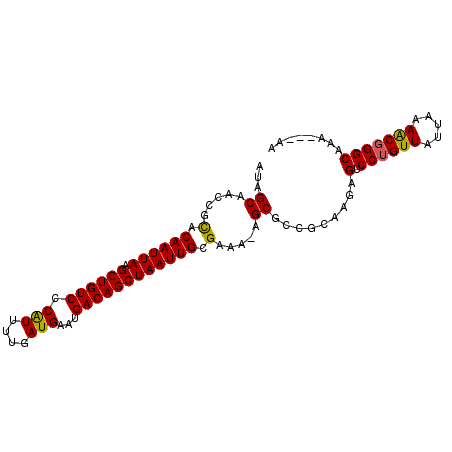
| Location | 14,437,877 – 14,437,973 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 93.22 |
| Mean single sequence MFE | -23.97 |
| Consensus MFE | -21.93 |
| Energy contribution | -21.60 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.55 |
| SVM RNA-class probability | 0.995152 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14437877 96 - 22407834 AUAGCAACCGCACAAUUAAGCUGUCCCAUUUUGAUGAAUGACAGCUAAUUGCGAAA-AGCGCCGCAAGAGUCCUGUUAUUAAAACGGGCAAAAAAAA ...((.....(.((((((.((((((.(((....)))...)))))))))))).)...-.))..(....).(.((((((.....)))))))........ ( -23.40) >DroSec_CAF1 28131 93 - 1 AUAGCAACCGCACAAUUAAGCUGUCCCAUGUUGAUGAAUGACAGCUAAUUGCGAAA-AGCGCCGCAAGAGUCCUGUUAUUAAAACGGGCAAA---AA ...((.....(.((((((.((((((.(((....)))...)))))))))))).)...-.))..(....).(.((((((.....)))))))...---.. ( -23.60) >DroSim_CAF1 30611 93 - 1 AUAGCAACCGCACAAUUAAGCUGUCCCAUUUUGAUGAAUGACAGCUAAUUGCGAAA-AGCGCCGCAAGAGUCCUGUUAUUAAAACGGGCAAA---AA ...((.....(.((((((.((((((.(((....)))...)))))))))))).)...-.))..(....).(.((((((.....)))))))...---.. ( -23.40) >DroEre_CAF1 30521 93 - 1 AUGGCAACCGCACAAUUAAGCUGUCCCAUUUUGAUGAAUGACAGCUAAUUGCGAAA-AGCGCCUGGAGAGUCCUGUUAUUAAAGCGGGCAAA---AA ..(((.....(.((((((.((((((.(((....)))...)))))))))))).)...-...)))......(.((((((.....)))))))...---.. ( -24.80) >DroYak_CAF1 30073 93 - 1 AUAGCAACCGCACAAUUAAGCUGUCCCGUUUUGAUGAAUGACAGCUAAUUGCGAAA-AGCGCCUGCAGAGUCCUGUUAUUAAAACGGGCAAA---AA ...(((..(((.((((((.((((((..((((....)))))))))))))))).....-.)))..)))...(.((((((.....)))))))...---.. ( -25.40) >DroAna_CAF1 20972 94 - 1 AUAGCCACCUUCCAAUUAAGCUGUCCCAUUUUGAUGAAUGACAGCUAAUUGCGAAAGAGCGCCUCGAGAGUCCUGUUAUUAAAGCCGGCAAA---AA ...(((.(.(((((((((.((((((.(((....)))...)))))))))))).))).)((((...(....)...)))).........)))...---.. ( -23.20) >consensus AUAGCAACCGCACAAUUAAGCUGUCCCAUUUUGAUGAAUGACAGCUAAUUGCGAAA_AGCGCCGCAAGAGUCCUGUUAUUAAAACGGGCAAA___AA ...((.....(.((((((.((((((.(((....)))...)))))))))))).).....)).........(.((((((.....)))))))........ (-21.93 = -21.60 + -0.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:34:12 2006