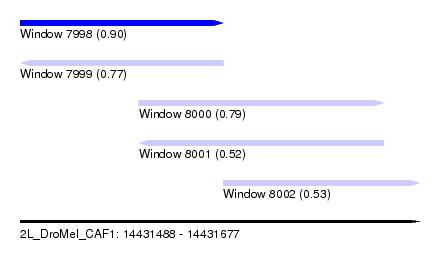
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,431,488 – 14,431,677 |
| Length | 189 |
| Max. P | 0.903598 |

| Location | 14,431,488 – 14,431,584 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 77.74 |
| Mean single sequence MFE | -34.17 |
| Consensus MFE | -16.82 |
| Energy contribution | -17.57 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.49 |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.903598 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14431488 96 + 22407834 CACCUAAAU-----------------GGGGGCGCUAAUCUCUGCAAC--AGAGAUUAACAGCCGA--GCAGACACAGAAACCAAUUUUGUGUGCCGGCCAACUGCCGGCCAGGCAGG ..(((....-----------------)))(((..(((((((((...)--))))))))...)))..--((..((((((((.....))))))))((((((.....))))))...))... ( -38.20) >DroPse_CAF1 30487 107 + 1 CAUCCAAAUAGAGGCGGGAGCGGGUUGGGGGCGCUAAUCUCUGCAAC--ACAGAUUAACUGCCGA--ACAGACACAGAAACCAAUUUUGUGUGCCAGUCAG------CCCAGGCAGC ...((........((....))(((((((.(((..((((((.((...)--).)))))).(((....--.)))((((((((.....)))))))))))..))))------))).)).... ( -32.30) >DroSec_CAF1 21771 93 + 1 CACCUAAAU-----------------GGGGGCGCUAAUCUCUGCAAC--AGAGAUUAACAGCCGA--GCAGACACAGAAACCAAUUUUGUGUGCCGGCCAACU---GGCCAGGCAGG ..(((....-----------------)))(((..(((((((((...)--))))))))...)))..--....((((((((.....))))))))(((((((....---)))).)))... ( -37.20) >DroEre_CAF1 23141 95 + 1 CACCUAAAU-----------------GGGGGCGCUAAUCUCUGCAAC--AGAGAUUAACAACCGGGCGCAGACACAGAAACCAAUUUUCUGUGCCAGCCAACU---GGCCAGGCAGG ..(((...(-----------------((((..(.(((((((((...)--)))))))).)..)).(((...(.((((((((.....)))))))))..)))....---..)))...))) ( -32.50) >DroAna_CAF1 14215 94 + 1 CGCCUAAAU-----------------GGGGGCGCUAAUCUCUGCAACACAGAGAUUAACAAACGAGAACAGACUCGGCAACCAAUUUUGGGUUCCAGCC---U---GGCCAGCCUGC .((((((((-----------------((..(((.(((((((((.....))))))))).)...((((......))))))..)))..)))))))....((.---.---((....)).)) ( -31.90) >DroPer_CAF1 17171 107 + 1 CAUCCAAAUAGAGGCGGGAGCGGGUUGGGGGCGCUAAUCUCUGCAAC--ACAGAUUAACAGCCGA--ACAGACACAGAAACCAAUUUUGUGUGCCAGUCAG------CCCAGGCAGC ...((........((....))(((((((.((((((....((((....--.)))).....)))...--....((((((((.....)))))))))))..))))------))).)).... ( -32.90) >consensus CACCUAAAU_________________GGGGGCGCUAAUCUCUGCAAC__AGAGAUUAACAGCCGA__ACAGACACAGAAACCAAUUUUGUGUGCCAGCCAA_U___GGCCAGGCAGC ...........................(((((..((((((((.......))))))))...)))........((((((((.....)))))))).)).(((............)))... (-16.82 = -17.57 + 0.75)

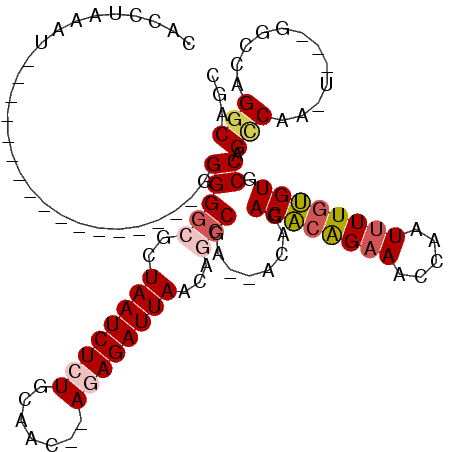
| Location | 14,431,488 – 14,431,584 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 77.74 |
| Mean single sequence MFE | -35.37 |
| Consensus MFE | -19.60 |
| Energy contribution | -20.55 |
| Covariance contribution | 0.95 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.768099 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14431488 96 - 22407834 CCUGCCUGGCCGGCAGUUGGCCGGCACACAAAAUUGGUUUCUGUGUCUGC--UCGGCUGUUAAUCUCU--GUUGCAGAGAUUAGCGCCCCC-----------------AUUUAGGUG ((((..(((..((((((((((.((((((.(((.....))).)))))).))--.)))))((((((((((--(...)))))))))))))).))-----------------)..)))).. ( -40.00) >DroPse_CAF1 30487 107 - 1 GCUGCCUGGG------CUGACUGGCACACAAAAUUGGUUUCUGUGUCUGU--UCGGCAGUUAAUCUGU--GUUGCAGAGAUUAGCGCCCCCAACCCGCUCCCGCCUCUAUUUGGAUG ((.(..((((------..(((.((((((.(((.....))).)))))).))--).(((.((((((((.(--(...)).)))))))))))))))..).)).....((.......))... ( -30.20) >DroSec_CAF1 21771 93 - 1 CCUGCCUGGCC---AGUUGGCCGGCACACAAAAUUGGUUUCUGUGUCUGC--UCGGCUGUUAAUCUCU--GUUGCAGAGAUUAGCGCCCCC-----------------AUUUAGGUG ...((((((((---....))))((((((.(((.....))).))))))...--..(((.((((((((((--(...))))))))))))))...-----------------....)))). ( -36.50) >DroEre_CAF1 23141 95 - 1 CCUGCCUGGCC---AGUUGGCUGGCACAGAAAAUUGGUUUCUGUGUCUGCGCCCGGUUGUUAAUCUCU--GUUGCAGAGAUUAGCGCCCCC-----------------AUUUAGGUG ((((..(((..---....(((.((((((((((.....))))))))))...))).((.(((((((((((--(...)))))))))))).))))-----------------)..)))).. ( -39.70) >DroAna_CAF1 14215 94 - 1 GCAGGCUGGCC---A---GGCUGGAACCCAAAAUUGGUUGCCGAGUCUGUUCUCGUUUGUUAAUCUCUGUGUUGCAGAGAUUAGCGCCCCC-----------------AUUUAGGCG ..((((.(..(---(---((((((((((.......)))).)).))))))..)..))))(((((((((((.....)))))))))))(((...-----------------.....))). ( -35.60) >DroPer_CAF1 17171 107 - 1 GCUGCCUGGG------CUGACUGGCACACAAAAUUGGUUUCUGUGUCUGU--UCGGCUGUUAAUCUGU--GUUGCAGAGAUUAGCGCCCCCAACCCGCUCCCGCCUCUAUUUGGAUG ((.(..((((------..(((.((((((.(((.....))).)))))).))--).(((.((((((((.(--(...)).)))))))))))))))..).)).....((.......))... ( -30.20) >consensus CCUGCCUGGCC___A_UUGGCUGGCACACAAAAUUGGUUUCUGUGUCUGC__UCGGCUGUUAAUCUCU__GUUGCAGAGAUUAGCGCCCCC_________________AUUUAGGUG ...(((.((((.......))))((((((.(((.....))).)))))).......))).((((((((((.......))))))))))................................ (-19.60 = -20.55 + 0.95)
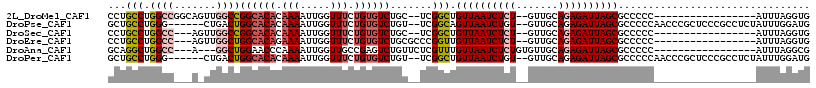

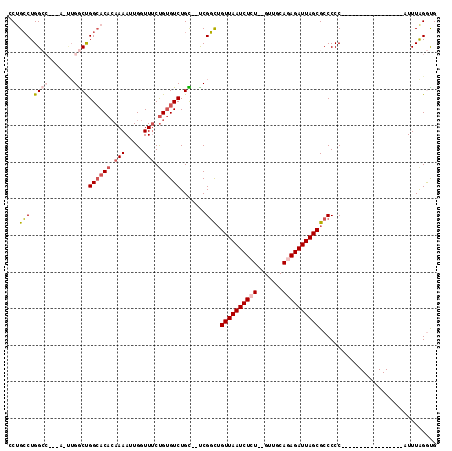
| Location | 14,431,544 – 14,431,660 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.10 |
| Mean single sequence MFE | -39.73 |
| Consensus MFE | -23.44 |
| Energy contribution | -23.64 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.785362 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
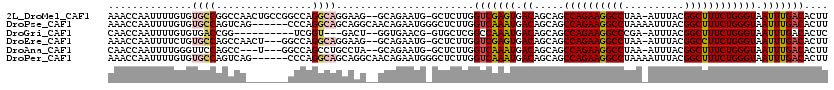
>2L_DroMel_CAF1 14431544 116 + 22407834 AAACCAAUUUUGUGUGCCGGCCAACUGCCGGCCAGGCAGGAAG--GCAGAAUG-GCUCUUGGUCGAGUGACAGCAGCCAGAAGGCCUAA-AUUUACGGCUUUCUGGGUAAUUUGACACUU ..(((((((((((.(((((((.....))))).)).)))))).(--((......-))).))))).(((((.(((.(.((((((((((...-......)))))))))).)...))).))))) ( -44.20) >DroPse_CAF1 30560 114 + 1 AAACCAAUUUUGUGUGCCAGUCAG------CCCAGGCAGCAGGCAACAGAAUGGGCUCUUGGUCAAAUGACAGCAGCCAGAAGGCCUAAAAUUUACGGCUUUCUGGGUAAUUUGACACUU ...(((.((((((.((((.((..(------(....)).)).))))))))))))).......(((((((.((.....((((((((((..........)))))))))))).))))))).... ( -39.60) >DroGri_CAF1 44659 103 + 1 CAACCAAUUUUGUGUGACCGG----------UCGGU---GACU--GGUGAACG-GUGCUCGGCCAAAUGACAGCAGCCAGAAGGCCCGA-AUUUACGGCUUUCUGGGUAAUUUGACACUC ...........((((.(..((----------(((((---.(((--(.....))-)))).)))))............((((((((((...-......))))))))))......).)))).. ( -33.30) >DroEre_CAF1 23199 113 + 1 AAACCAAUUUUCUGUGCCAGCCAACU---GGCCAGGCAGGAAG--GCAGAAUG-GCUCUUGGUCGAGUGACAGCAGCCAGAAGGCCUAA-AUUUACGGCCUUCUGGGUAAUUUGACACUU ..(((((.((((((((((((....))---)))...)))))))(--((......-))).))))).(((((.(((.(.((((((((((...-......)))))))))).)...))).))))) ( -43.20) >DroAna_CAF1 14275 110 + 1 CAACCAAUUUUGGGUUCCAGCC---U---GGCCAGCCUGCCUA--GCAGAAUG-GCUCUUGGUCAAAUGACAGCAGCCAGAAGGCCUAA-AUUUACGGCUUUCUGGGUAAUUUGACACUU ....(((....((((....)))---)---(((((..((((...--))))..))-))).)))(((((((.((.....((((((((((...-......)))))))))))).))))))).... ( -38.50) >DroPer_CAF1 17244 114 + 1 AAACCAAUUUUGUGUGCCAGUCAG------CCCAGGCAGCAGGCAACAGAAUGGGCUCUUGGUCAAAUGACAGCAGCCAGAAGGCCUAAAAUUUACGGCUUUCUGGGUAAUUUGACACUU ...(((.((((((.((((.((..(------(....)).)).))))))))))))).......(((((((.((.....((((((((((..........)))))))))))).))))))).... ( -39.60) >consensus AAACCAAUUUUGUGUGCCAGCCA_______GCCAGGCAGCAAG__GCAGAAUG_GCUCUUGGUCAAAUGACAGCAGCCAGAAGGCCUAA_AUUUACGGCUUUCUGGGUAAUUUGACACUU ..............((((................)))).......................(((((((.((.....((((((((((..........)))))))))))).))))))).... (-23.44 = -23.64 + 0.20)


| Location | 14,431,544 – 14,431,660 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.10 |
| Mean single sequence MFE | -35.98 |
| Consensus MFE | -21.30 |
| Energy contribution | -21.36 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.59 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.522775 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
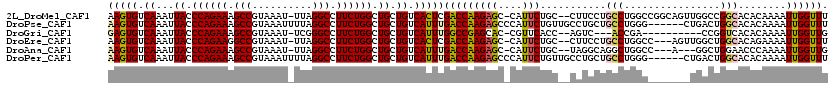
>2L_DroMel_CAF1 14431544 116 - 22407834 AAGUGUCAAAUUACCCAGAAAGCCGUAAAU-UUAGGCCUUCUGGCUGCUGUCACUCGACCAAGAGC-CAUUCUGC--CUUCCUGCCUGGCCGGCAGUUGGCCGGCACACAAAAUUGGUUU ..(((((.......((((((.(((......-...))).))))))((...(((....)))..)).((-((..((((--(..((.....))..))))).)))).)))))............. ( -39.30) >DroPse_CAF1 30560 114 - 1 AAGUGUCAAAUUACCCAGAAAGCCGUAAAUUUUAGGCCUUCUGGCUGCUGUCAUUUGACCAAGAGCCCAUUCUGUUGCCUGCUGCCUGGG------CUGACUGGCACACAAAAUUGGUUU ..(.(((((((.((((((((.(((..........))).)))))).....)).))))))))..(((((.(((.(((((((..(.((....)------).)...)))).))).))).))))) ( -36.30) >DroGri_CAF1 44659 103 - 1 GAGUGUCAAAUUACCCAGAAAGCCGUAAAU-UCGGGCCUUCUGGCUGCUGUCAUUUGGCCGAGCAC-CGUUCACC--AGUC---ACCGA----------CCGGUCACACAAAAUUGGUUG ..(.(((((((.((((((((.(((......-...))).)))))).....)).))))))))((((..-.))))...--....---..(((----------(((((........)))))))) ( -26.70) >DroEre_CAF1 23199 113 - 1 AAGUGUCAAAUUACCCAGAAGGCCGUAAAU-UUAGGCCUUCUGGCUGCUGUCACUCGACCAAGAGC-CAUUCUGC--CUUCCUGCCUGGCC---AGUUGGCUGGCACAGAAAAUUGGUUU .((((.((...((.((((((((((......-...)))))))))).)).)).)))).......((((-(((((((.--.....((((.((((---....)))))))))))))...)))))) ( -43.00) >DroAna_CAF1 14275 110 - 1 AAGUGUCAAAUUACCCAGAAAGCCGUAAAU-UUAGGCCUUCUGGCUGCUGUCAUUUGACCAAGAGC-CAUUCUGC--UAGGCAGGCUGGCC---A---GGCUGGAACCCAAAAUUGGUUG (((((.((...((.((((((.(((......-...))).)))))).)).)).)))))(((((((.((-((..((((--...))))..)))))---.---...(((...)))...)))))). ( -34.30) >DroPer_CAF1 17244 114 - 1 AAGUGUCAAAUUACCCAGAAAGCCGUAAAUUUUAGGCCUUCUGGCUGCUGUCAUUUGACCAAGAGCCCAUUCUGUUGCCUGCUGCCUGGG------CUGACUGGCACACAAAAUUGGUUU ..(.(((((((.((((((((.(((..........))).)))))).....)).))))))))..(((((.(((.(((((((..(.((....)------).)...)))).))).))).))))) ( -36.30) >consensus AAGUGUCAAAUUACCCAGAAAGCCGUAAAU_UUAGGCCUUCUGGCUGCUGUCAUUUGACCAAGAGC_CAUUCUGC__CCUCCUGCCUGGC_______UGGCUGGCACACAAAAUUGGUUU (((((.((...((.((((((.(((..........))).)))))).)).)).)))))(((((((((....)))...........(((................)))........)))))). (-21.30 = -21.36 + 0.06)

| Location | 14,431,584 – 14,431,677 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 84.30 |
| Mean single sequence MFE | -27.17 |
| Consensus MFE | -22.82 |
| Energy contribution | -22.40 |
| Covariance contribution | -0.42 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.84 |
| SVM decision value | -0.00 |
| SVM RNA-class probability | 0.531088 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
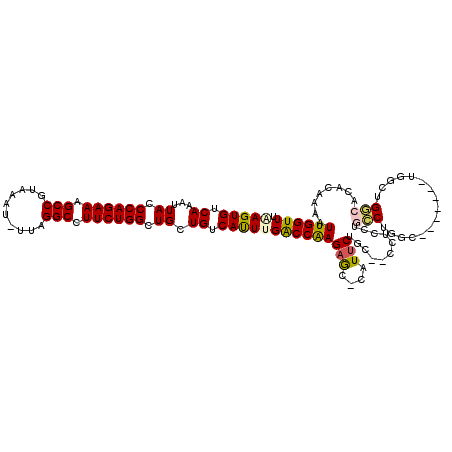
>2L_DroMel_CAF1 14431584 93 + 22407834 AAG--GCAGAAUG-GCUCUUGGUCGAGUGACAGCAGCCAGAAGGCCUAA-AUUUACGGCUUUCUGGGUAAUUUGACACUU-CGGACAG---AAUAAA-C-GAG ..(--((......-))).(((..((((((.(((.(.((((((((((...-......)))))))))).)...))).))).)-))..)))---......-.-... ( -27.40) >DroPse_CAF1 30594 98 + 1 AGGCAACAGAAUGGGCUCUUGGUCAAAUGACAGCAGCCAGAAGGCCUAAAAUUUACGGCUUUCUGGGUAAUUUGACACUUUUCGACAG---AAUAUA-C-GAG .(....)..........(((((((((((.((.....((((((((((..........)))))))))))).)))))))....(((....)---))....-)-))) ( -26.40) >DroGri_CAF1 44686 95 + 1 ACU--GGUGAACG-GUGCUCGGCCAAAUGACAGCAGCCAGAAGGCCCGA-AUUUACGGCUUUCUGGGUAAUUUGACACUC-UGGACAGCAGCACAAUCCC--- ...--((.((...-((((((..(((..((.(((.(.((((((((((...-......)))))))))).)...))).))...-)))...).)))))..))))--- ( -28.90) >DroEre_CAF1 23236 93 + 1 AAG--GCAGAAUG-GCUCUUGGUCGAGUGACAGCAGCCAGAAGGCCUAA-AUUUACGGCCUUCUGGGUAAUUUGACACUU-UGGACAG---AAUGAA-C-GAG (((--((......-)).))).((((((((.(((.(.((((((((((...-......)))))))))).)...))).)))))-..)))..---......-.-... ( -29.50) >DroAna_CAF1 14309 93 + 1 CUA--GCAGAAUG-GCUCUUGGUCAAAUGACAGCAGCCAGAAGGCCUAA-AUUUACGGCUUUCUGGGUAAUUUGACACUU-UGGACAG---AAUAAU-C-GAG ..(--((......-)))(((((((((((.((.....((((((((((...-......)))))))))))).))))))).((.-.....))---......-)-))) ( -24.40) >DroPer_CAF1 17278 98 + 1 AGGCAACAGAAUGGGCUCUUGGUCAAAUGACAGCAGCCAGAAGGCCUAAAAUUUACGGCUUUCUGGGUAAUUUGACACUUUUCGACAG---AAUAUA-C-GAG .(....)..........(((((((((((.((.....((((((((((..........)))))))))))).)))))))....(((....)---))....-)-))) ( -26.40) >consensus AAG__GCAGAAUG_GCUCUUGGUCAAAUGACAGCAGCCAGAAGGCCUAA_AUUUACGGCUUUCUGGGUAAUUUGACACUU_UGGACAG___AAUAAA_C_GAG .....................(((((((.((.....((((((((((..........)))))))))))).)))))))........................... (-22.82 = -22.40 + -0.42)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:34:09 2006