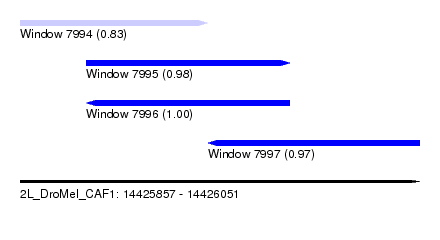
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,425,857 – 14,426,051 |
| Length | 194 |
| Max. P | 0.999807 |

| Location | 14,425,857 – 14,425,948 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 77.68 |
| Mean single sequence MFE | -31.95 |
| Consensus MFE | -18.69 |
| Energy contribution | -19.31 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.827663 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14425857 91 + 22407834 ----CCGCCAGUCCGCCUGUCAGCAGUCAAUCGGCUACC-GUCUGUCAAUGUCGUGUCGCCUUCAUCAAAGGCCACAUU-------CGAGUUGACAAUGGG---GA ----(((((.((..(.(((....))).).)).)))..((-((.(((((((...((((.(((((.....))))).)))).-------...))))))))))))---). ( -29.90) >DroVir_CAF1 33041 100 + 1 AGUAGCUGCUGGCUGGCUGUCAGAAGUCAAUCGGCUGCC-GUCUGUCAAUGUCGUGUCGCCUUCAUCAAAGGCGAAACUCGAGCGUUGAGUUGGCAAUGGA----- .......((((..(((((......)))))..))))((((-(....(((((((((..(((((((.....)))))))....))).))))))..))))).....----- ( -38.60) >DroEre_CAF1 17631 91 + 1 ----CCGCCAGUUGGCCUGUCAGCAGUCAAUCGGCUGCC-GUCUGUCAAUGUCGUGUCGCCUUCAUCAAAGGCCACAUU-------CGAGUUGACAAUGGG---GA ----(((((.((((((.((....)))))))).)))..((-((.(((((((...((((.(((((.....))))).)))).-------...))))))))))))---). ( -35.70) >DroWil_CAF1 115217 73 + 1 ------------GCUGCUGUCAGUAGUCAAUCGGCUACC-GUCUGUCAAUGUCGUGUCGCCUUCAUCAAAGGC---------------AAUUGACAAUGGC----- ------------(((..(((((((((((....)))))).-..................(((((.....)))))---------------...)))))..)))----- ( -21.90) >DroAna_CAF1 8474 91 + 1 ----CACCCGGACGGCCUGUCAGUAGUCAAUCGGCUGCC-GUCUGUCAACGUCGUGUCGCCUUCAUCAAAGGCCGCAUU-------CGAGUUGACAAUGGG---GA ----..((((((((((..(((.((.....)).))).)))-))))((((((.((((((.(((((.....))))).)))..-------)))))))))...)))---.. ( -38.80) >DroPer_CAF1 5262 95 + 1 ----CCUUCUGUCUGCCUGUCAGUAGUCAAUCGGCUACCGGUCUGUCAAUGUCGUGUCGCCUUCAUCAAAGGCCGCUUU-------CAACUUGACAAUGGGAUGGA ----...((((((((((.....((((((....)))))).))).((((((.((.(.((.(((((.....))))).))...-------).))))))))...))))))) ( -26.80) >consensus ____CCGCCAGUCCGCCUGUCAGUAGUCAAUCGGCUACC_GUCUGUCAAUGUCGUGUCGCCUUCAUCAAAGGCCACAUU_______CGAGUUGACAAUGGG___GA ...............(((((..((((((....)))))).....(((((((...((((.(((((.....))))).))))...........))))))))))))..... (-18.69 = -19.31 + 0.61)
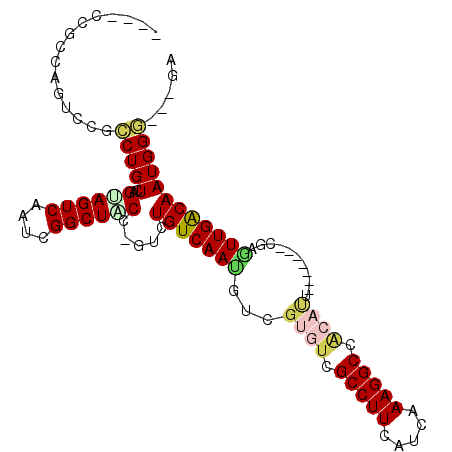
| Location | 14,425,889 – 14,425,988 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 92.12 |
| Mean single sequence MFE | -29.40 |
| Consensus MFE | -26.93 |
| Energy contribution | -26.70 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.976174 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14425889 99 + 22407834 ACCGUCUGUCAAUGUCGUGUCGCCUUCAUCAAAGGCCACAUUCGAGUUGACAAUGGGGAUAUUGGUUCUGUUUUGUAAUCCGCAAAGGAUACGGAUUUU .((((.(((((((...((((.(((((.....))))).))))....)))))))))))((((....))))...((((((.(((.....))))))))).... ( -29.00) >DroSec_CAF1 16385 99 + 1 ACCGUCUGUCAAUGUCGUGUCGCCUUCAUCAAAGGCCACAUUCGAGUUGACAAUGGGGAUAUUGGUUCUGUUUUGUAAUCCGCAAAGGAUACGGAUUUU .((((.(((((((...((((.(((((.....))))).))))....)))))))))))((((....))))...((((((.(((.....))))))))).... ( -29.00) >DroSim_CAF1 18057 99 + 1 ACCGUCUGUCAAUGUCGUGUCGCCUUCAUCAAAGGCCACAUUCGAGUUGACAAUGGGGAUAUUGGUUCUGUUUUGUAAUCCGCAAAGGAUACGGAUUUU .((((.(((((((...((((.(((((.....))))).))))....)))))))))))((((....))))...((((((.(((.....))))))))).... ( -29.00) >DroEre_CAF1 17663 99 + 1 GCCGUCUGUCAAUGUCGUGUCGCCUUCAUCAAAGGCCACAUUCGAGUUGACAAUGGGGAUAUUGGUUCUGUUUGGUAAUCCGCAAAGGAUACGGAUUUU .((((.(((((((...((((.(((((.....))))).))))....))))))).((.((((((..(......)..)).)))).))......))))..... ( -30.50) >DroYak_CAF1 17893 99 + 1 ACCGUCUGUCAAUGUCGUGUCGCCUUCAUCAAAGGCCACAUUCGAGUUGACAAUGGGGAUAUUGGUUCUGUUUUGUAAUCCGCAAAGGAUACGGAUUUU .((((.(((((((...((((.(((((.....))))).))))....)))))))))))((((....))))...((((((.(((.....))))))))).... ( -29.00) >DroAna_CAF1 8506 80 + 1 GCCGUCUGUCAACGUCGUGUCGCCUUCAUCAAAGGCCGCAUUCGAGUUGACAAUGGGGA-------------------UCCGCAAAGGAUACGGAUUUU .((((.(((((((.((((((.(((((.....))))).)))..))))))))))))))(((-------------------((((.........))))))). ( -29.90) >consensus ACCGUCUGUCAAUGUCGUGUCGCCUUCAUCAAAGGCCACAUUCGAGUUGACAAUGGGGAUAUUGGUUCUGUUUUGUAAUCCGCAAAGGAUACGGAUUUU .((((.(((((((.((((((.(((((.....))))).)))..)))))))))).((.((((.................)))).))......))))..... (-26.93 = -26.70 + -0.24)

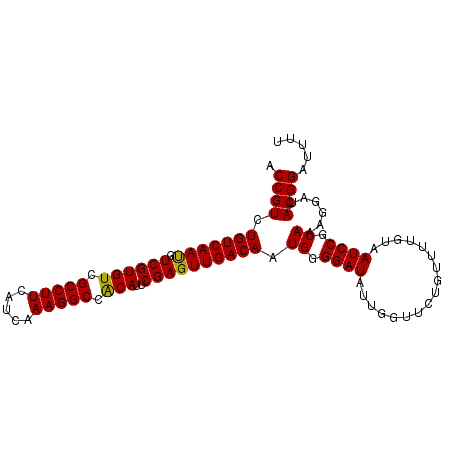
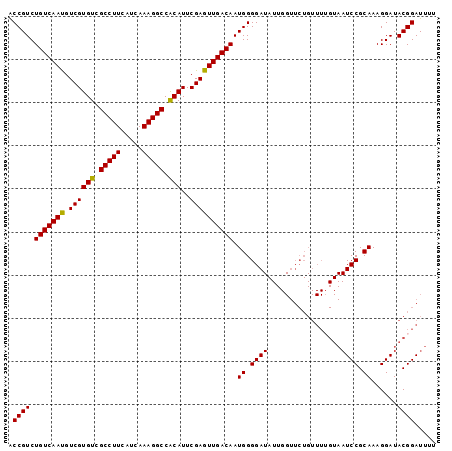
| Location | 14,425,889 – 14,425,988 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 92.12 |
| Mean single sequence MFE | -28.90 |
| Consensus MFE | -27.12 |
| Energy contribution | -27.62 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.88 |
| Structure conservation index | 0.94 |
| SVM decision value | 4.13 |
| SVM RNA-class probability | 0.999807 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14425889 99 - 22407834 AAAAUCCGUAUCCUUUGCGGAUUACAAAACAGAACCAAUAUCCCCAUUGUCAACUCGAAUGUGGCCUUUGAUGAAGGCGACACGACAUUGACAGACGGU ..((((((((.....))))))))...................((..(((((((.(((..(((.((((((...)))))).))))))..)))))))..)). ( -29.20) >DroSec_CAF1 16385 99 - 1 AAAAUCCGUAUCCUUUGCGGAUUACAAAACAGAACCAAUAUCCCCAUUGUCAACUCGAAUGUGGCCUUUGAUGAAGGCGACACGACAUUGACAGACGGU ..((((((((.....))))))))...................((..(((((((.(((..(((.((((((...)))))).))))))..)))))))..)). ( -29.20) >DroSim_CAF1 18057 99 - 1 AAAAUCCGUAUCCUUUGCGGAUUACAAAACAGAACCAAUAUCCCCAUUGUCAACUCGAAUGUGGCCUUUGAUGAAGGCGACACGACAUUGACAGACGGU ..((((((((.....))))))))...................((..(((((((.(((..(((.((((((...)))))).))))))..)))))))..)). ( -29.20) >DroEre_CAF1 17663 99 - 1 AAAAUCCGUAUCCUUUGCGGAUUACCAAACAGAACCAAUAUCCCCAUUGUCAACUCGAAUGUGGCCUUUGAUGAAGGCGACACGACAUUGACAGACGGC ..((((((((.....))))))))...................((..(((((((.(((..(((.((((((...)))))).))))))..)))))))..)). ( -29.00) >DroYak_CAF1 17893 99 - 1 AAAAUCCGUAUCCUUUGCGGAUUACAAAACAGAACCAAUAUCCCCAUUGUCAACUCGAAUGUGGCCUUUGAUGAAGGCGACACGACAUUGACAGACGGU ..((((((((.....))))))))...................((..(((((((.(((..(((.((((((...)))))).))))))..)))))))..)). ( -29.20) >DroAna_CAF1 8506 80 - 1 AAAAUCCGUAUCCUUUGCGGA-------------------UCCCCAUUGUCAACUCGAAUGCGGCCUUUGAUGAAGGCGACACGACGUUGACAGACGGC ...(((((((.....))))))-------------------).((..(((((((((((..((..((((((...))))))..))))).))))))))..)). ( -27.60) >consensus AAAAUCCGUAUCCUUUGCGGAUUACAAAACAGAACCAAUAUCCCCAUUGUCAACUCGAAUGUGGCCUUUGAUGAAGGCGACACGACAUUGACAGACGGU ..((((((((.....))))))))...................((..(((((((.(((..(((.((((((...)))))).))))))..)))))))..)). (-27.12 = -27.62 + 0.50)

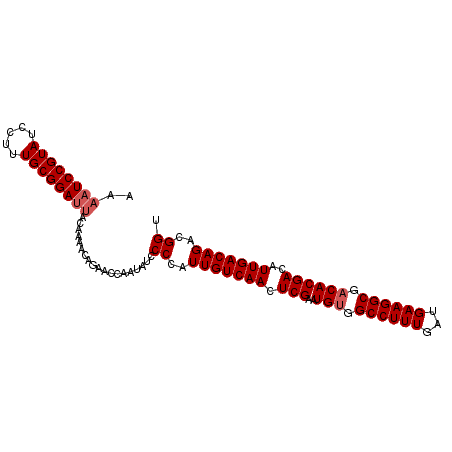
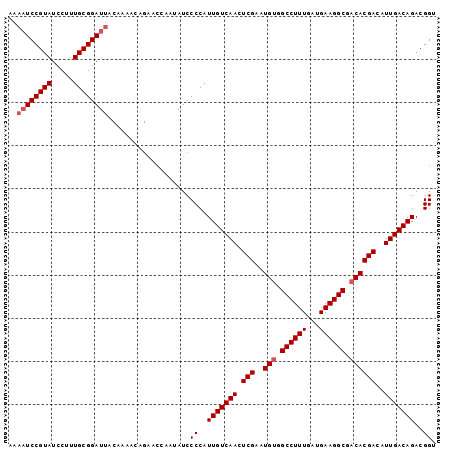
| Location | 14,425,948 – 14,426,051 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 84.34 |
| Mean single sequence MFE | -20.22 |
| Consensus MFE | -17.96 |
| Energy contribution | -17.80 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.71 |
| SVM RNA-class probability | 0.973446 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14425948 103 - 22407834 AAUUGCAUUCAAGUAUGUUCCAUCCAGCCACGUAGCUCUUCUUGCC-AGACUUGAGCACUUUCAAAAAUCCGUAUCCUUUGCGGAUUACAAAACAGAACCAAUA ...(((..((((((.((........(((......)))........)-).)))))))))........((((((((.....))))))))................. ( -19.49) >DroSec_CAF1 16444 104 - 1 AAUUGCAUUCAACUAUGUUCCCUCCAGCCACGUAGCUCUUCUUGCCAAGACUUGAGCACUUUCGAAAAUCCGUAUCCUUUGCGGAUUACAAAACAGAACCAAUA ................((((...........((.((((.(((.....)))...)))))).......((((((((.....))))))))........))))..... ( -18.70) >DroSim_CAF1 18116 104 - 1 AAUUGCAUUCAACUAUGUUCCCUCCAGCCACGUAGCUCUUCUGGCCAAGACUUGAGCACUUUCGAAAAUCCGUAUCCUUUGCGGAUUACAAAACAGAACCAAUA ................((((...........((.((((.(((.....)))...)))))).......((((((((.....))))))))........))))..... ( -19.80) >DroEre_CAF1 17722 80 - 1 AAUUGCAUUC-----------------------AGCUCUUCUUGCC-GGAGCUGAGCACUUUCGAAAAUCCGUAUCCUUUGCGGAUUACCAAACAGAACCAAUA ...(((..((-----------------------((((((.......-)))))))))))........((((((((.....))))))))................. ( -22.80) >DroYak_CAF1 17952 88 - 1 AAUUGCAUUCAACUAUG---------------CAGCUCUUCUUGCC-AGACUUGAGCACUUUCGAAAAUCCGUAUCCUUUGCGGAUUACAAAACAGAACCAAUA ...(((((......)))---------------))((((.(((....-)))...)))).........((((((((.....))))))))................. ( -20.30) >consensus AAUUGCAUUCAACUAUGUUCC_UCCAGCCACGUAGCUCUUCUUGCC_AGACUUGAGCACUUUCGAAAAUCCGUAUCCUUUGCGGAUUACAAAACAGAACCAAUA .((((..(((........................((((.(((.....)))...)))).........((((((((.....))))))))........))).)))). (-17.96 = -17.80 + -0.16)

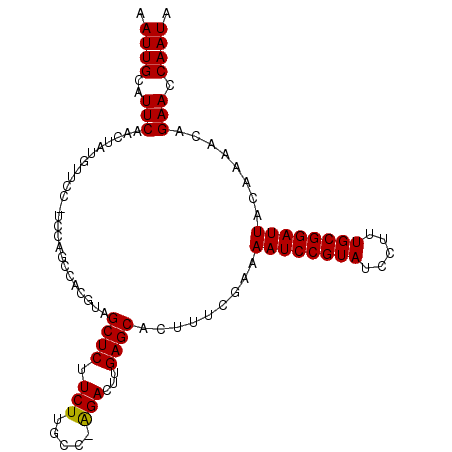
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:34:05 2006