
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,424,967 – 14,425,094 |
| Length | 127 |
| Max. P | 0.988348 |

| Location | 14,424,967 – 14,425,068 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 85.81 |
| Mean single sequence MFE | -36.42 |
| Consensus MFE | -29.89 |
| Energy contribution | -29.58 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.676687 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14424967 101 + 22407834 GGCUCCACCGUGAUGACCGUGGUGUUCGUGGCCAUGGAAAGGAGCUGCCCGAAUAGCCGCAAUGGCCAAGAGGGCAAAGGCGAUGAAAAACUAAGGAAAAU ((((((.(......).(((((((.......)))))))...))))))((((.....(((.....))).....)))).......................... ( -30.30) >DroSec_CAF1 15498 101 + 1 GGCUCCACCAUGAUGGCCGUGGUGUUCGUGGCCGUGGAAAGGAGCUGCCCGGAUAGCCGCAAUGGCCAAGAGGGCAAAGGCGAUGCAAAACUAAGGGAAAU ((((((.((((...(((((((.....)))))))))))...)))))).(((((....))((....(((.....)))....)).............))).... ( -35.00) >DroSim_CAF1 17169 101 + 1 GGCUCCACCAUGAUGGCCGUGGUGUUCGUGGCCAUGGAAAGGAGCUGCCCGGAUAGCCGCAAUGGCCAAGAGGGCAAAGGCGAUGAAAAACUAAGGGAAAU ((((((.(((...((((((((.....)))))))))))...)))))).(((((....))((....(((.....)))....)).............))).... ( -35.20) >DroEre_CAF1 16761 101 + 1 GGCACCGCCAUGGUGGCCAUGGUGUUCGUGGCCGUGGAAAGGUGCUGCCCGGAUAGCCGCAGUUGCCAGGAGGGCAAAGGCGAUGCAAAACUGAGGGAUAU ((((((.((((...(((((((.....)))))))))))...)))))).(((((....))((..(((((.....)))))..)).............))).... ( -43.70) >DroYak_CAF1 16990 101 + 1 GGCGCCACCAUGAUGGCCAUGGUGUUCGUGGCCGUGGAAAGGAGCUGCCCGGAUAGCCGCAAUGGCCAAGAGGGCAAAGGCGAUGAAGAACUAGGGGAAAU ..((((.((((...(((((((.....)))))))))))........(((((.....(((.....))).....)))))..))))................... ( -36.20) >DroAna_CAF1 7643 98 + 1 GGCACCUCCA---UGGCUGUGGUGUUCGUGGCCAUGGAAAGGAGCUGCCCGAAUGGCAGCGACUGCCACCAGAGCGAAUACUACGAAAAACUGAGAAAAGA (((...((((---((((..((.....))..)))))))).((..((((((.....))))))..)))))..(((..((.......)).....)))........ ( -38.10) >consensus GGCUCCACCAUGAUGGCCGUGGUGUUCGUGGCCAUGGAAAGGAGCUGCCCGGAUAGCCGCAAUGGCCAAGAGGGCAAAGGCGAUGAAAAACUAAGGGAAAU ((((((.(((...((((((((.....)))))))))))...))))))((((.....(((.....))).....)))).......................... (-29.89 = -29.58 + -0.30)



| Location | 14,424,967 – 14,425,068 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 85.81 |
| Mean single sequence MFE | -33.82 |
| Consensus MFE | -27.97 |
| Energy contribution | -28.17 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.905695 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14424967 101 - 22407834 AUUUUCCUUAGUUUUUCAUCGCCUUUGCCCUCUUGGCCAUUGCGGCUAUUCGGGCAGCUCCUUUCCAUGGCCACGAACACCACGGUCAUCACGGUGGAGCC .........................(((((...(((((.....)))))...)))))(((((...((((((((...........))))))...)).))))). ( -32.20) >DroSec_CAF1 15498 101 - 1 AUUUCCCUUAGUUUUGCAUCGCCUUUGCCCUCUUGGCCAUUGCGGCUAUCCGGGCAGCUCCUUUCCACGGCCACGAACACCACGGCCAUCAUGGUGGAGCC .........................(((((...(((((.....)))))...)))))(((((...(((.((((...........))))....))).))))). ( -33.50) >DroSim_CAF1 17169 101 - 1 AUUUCCCUUAGUUUUUCAUCGCCUUUGCCCUCUUGGCCAUUGCGGCUAUCCGGGCAGCUCCUUUCCAUGGCCACGAACACCACGGCCAUCAUGGUGGAGCC .........................(((((...(((((.....)))))...)))))(((((...((((((((...........)))))...))).))))). ( -35.40) >DroEre_CAF1 16761 101 - 1 AUAUCCCUCAGUUUUGCAUCGCCUUUGCCCUCCUGGCAACUGCGGCUAUCCGGGCAGCACCUUUCCACGGCCACGAACACCAUGGCCACCAUGGCGGUGCC ...............((((((((.((((((...((((.......))))...))))))...........(((((.(.....).))))).....)))))))). ( -36.00) >DroYak_CAF1 16990 101 - 1 AUUUCCCCUAGUUCUUCAUCGCCUUUGCCCUCUUGGCCAUUGCGGCUAUCCGGGCAGCUCCUUUCCACGGCCACGAACACCAUGGCCAUCAUGGUGGCGCC ...................((((.((((((...(((((.....)))))...)))))).......(((.(((((.(.....).)))))....))).)))).. ( -31.80) >DroAna_CAF1 7643 98 - 1 UCUUUUCUCAGUUUUUCGUAGUAUUCGCUCUGGUGGCAGUCGCUGCCAUUCGGGCAGCUCCUUUCCAUGGCCACGAACACCACAGCCA---UGGAGGUGCC ..........................((((..(((((((...)))))))..)))).((.((..((((((((.............))))---)))))).)). ( -34.02) >consensus AUUUCCCUUAGUUUUUCAUCGCCUUUGCCCUCUUGGCCAUUGCGGCUAUCCGGGCAGCUCCUUUCCACGGCCACGAACACCACGGCCAUCAUGGUGGAGCC .........................(((((...(((((.....)))))...)))))(((((...(((.((((...........))))....))).))))). (-27.97 = -28.17 + 0.20)

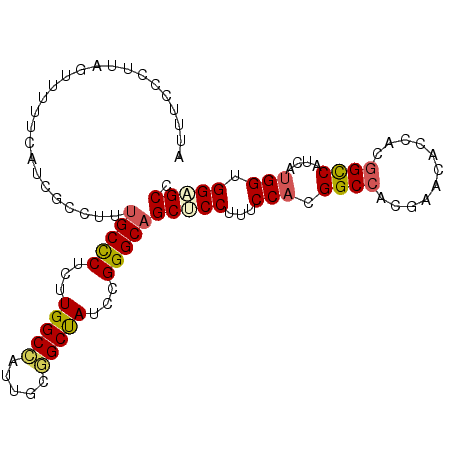
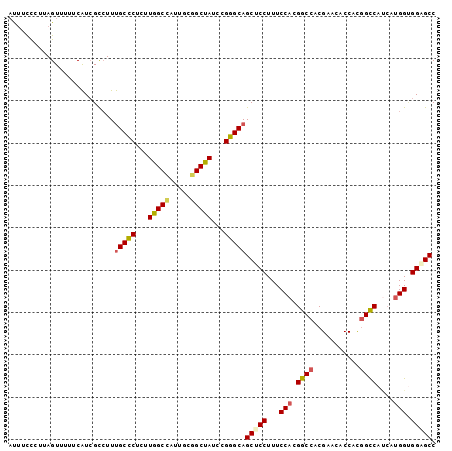
| Location | 14,425,004 – 14,425,094 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 76.53 |
| Mean single sequence MFE | -23.00 |
| Consensus MFE | -11.46 |
| Energy contribution | -11.60 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.667032 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14425004 90 + 22407834 AAAGGAGCUGCCCGAAUAGCCGCAAUGGCCAAGAGGGCAAAGGCGAUGAAAAACUAAGGAAAAUCGAGGUGAAUAAUAAUUUGC-UUUAGU ......(((((((.....(((.....))).....))))...))).....................((((..(((....)))..)-)))... ( -19.80) >DroSec_CAF1 15535 90 + 1 AAAGGAGCUGCCCGGAUAGCCGCAAUGGCCAAGAGGGCAAAGGCGAUGCAAAACUAAGGGAAAUCAAGGAAAAUAAUGAAUUCC-UUUAGU ......(((((((.....(((.....))).....))))...)))........((((((((((.(((..........))).))))-)))))) ( -26.70) >DroSim_CAF1 17206 90 + 1 AAAGGAGCUGCCCGGAUAGCCGCAAUGGCCAAGAGGGCAAAGGCGAUGAAAAACUAAGGGAAAUCCAUGGAAAUAAUGAACUCC-UUUAGU ......(((((((.....(((.....))).....))))...)))........(((((((((....(((.......)))...)))-)))))) ( -24.50) >DroEre_CAF1 16798 91 + 1 AAAGGUGCUGCCCGGAUAGCCGCAGUUGCCAGGAGGGCAAAGGCGAUGCAAAACUGAGGGAUAUCGAGGGAAAUAAUAAAUGCCAUUCGGC ..........(((.((((.((((..(((((.....)))))..))....((....))..)).))))..)))...........(((....))) ( -23.20) >DroYak_CAF1 17027 90 + 1 AAAGGAGCUGCCCGGAUAGCCGCAAUGGCCAAGAGGGCAAAGGCGAUGAAGAACUAGGGGAAAUCGAUGGAGGUAAUUACUUCC-UUUGGU ......(((((((.....(((.....))).....))))...)))..................(((((.((((((....))))))-.))))) ( -24.30) >DroAna_CAF1 7677 82 + 1 AAAGGAGCUGCCCGAAUGGCAGCGACUGCCACCAGAGCGAAUACUACGAAAAACUGAGAAAAGA-AAAGGA-------UUUUGU-AAGGAU ..((..((((((.....))))))..))(((....).))......(((((((..((.........-..))..-------))))))-)..... ( -19.50) >consensus AAAGGAGCUGCCCGGAUAGCCGCAAUGGCCAAGAGGGCAAAGGCGAUGAAAAACUAAGGGAAAUCGAGGGAAAUAAUAAAUUCC_UUUAGU ......(((((((.....(((.....))).....)))))...))............................................... (-11.46 = -11.60 + 0.14)

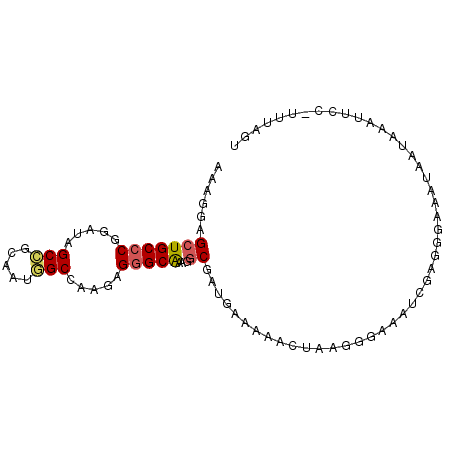
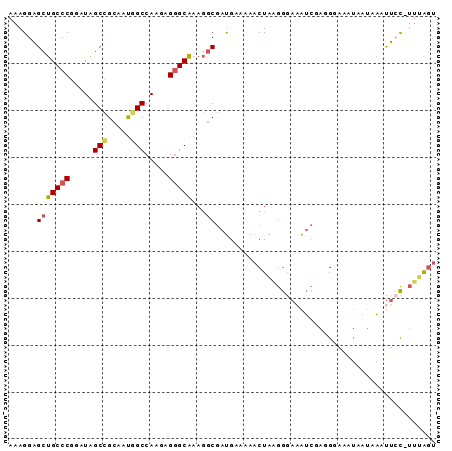
| Location | 14,425,004 – 14,425,094 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 76.53 |
| Mean single sequence MFE | -21.98 |
| Consensus MFE | -15.39 |
| Energy contribution | -15.08 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.59 |
| Structure conservation index | 0.70 |
| SVM decision value | 2.12 |
| SVM RNA-class probability | 0.988348 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14425004 90 - 22407834 ACUAAA-GCAAAUUAUUAUUCACCUCGAUUUUCCUUAGUUUUUCAUCGCCUUUGCCCUCUUGGCCAUUGCGGCUAUUCGGGCAGCUCCUUU (((((.-(.(((((............))))).).)))))........((...(((((...(((((.....)))))...)))))))...... ( -18.70) >DroSec_CAF1 15535 90 - 1 ACUAAA-GGAAUUCAUUAUUUUCCUUGAUUUCCCUUAGUUUUGCAUCGCCUUUGCCCUCUUGGCCAUUGCGGCUAUCCGGGCAGCUCCUUU (((((.-((((.(((..........))).)))).)))))........((...(((((...(((((.....)))))...)))))))...... ( -26.50) >DroSim_CAF1 17206 90 - 1 ACUAAA-GGAGUUCAUUAUUUCCAUGGAUUUCCCUUAGUUUUUCAUCGCCUUUGCCCUCUUGGCCAUUGCGGCUAUCCGGGCAGCUCCUUU (((((.-(((((((((.......))))).)))).)))))........((...(((((...(((((.....)))))...)))))))...... ( -26.10) >DroEre_CAF1 16798 91 - 1 GCCGAAUGGCAUUUAUUAUUUCCCUCGAUAUCCCUCAGUUUUGCAUCGCCUUUGCCCUCCUGGCAACUGCGGCUAUCCGGGCAGCACCUUU .(((.(((((................((......))..........(((..(((((.....)))))..)))))))).)))........... ( -20.40) >DroYak_CAF1 17027 90 - 1 ACCAAA-GGAAGUAAUUACCUCCAUCGAUUUCCCCUAGUUCUUCAUCGCCUUUGCCCUCUUGGCCAUUGCGGCUAUCCGGGCAGCUCCUUU ...(((-(((.((............((((...............))))....(((((...(((((.....)))))...))))))))))))) ( -21.96) >DroAna_CAF1 7677 82 - 1 AUCCUU-ACAAAA-------UCCUUU-UCUUUUCUCAGUUUUUCGUAGUAUUCGCUCUGGUGGCAGUCGCUGCCAUUCGGGCAGCUCCUUU ....((-((.(((-------..((..-.........))..))).)))).....((((..(((((((...)))))))..))))......... ( -18.20) >consensus ACCAAA_GGAAUUCAUUAUUUCCCUCGAUUUCCCUUAGUUUUUCAUCGCCUUUGCCCUCUUGGCCAUUGCGGCUAUCCGGGCAGCUCCUUU ...............................................((...(((((...(((((.....)))))...)))))))...... (-15.39 = -15.08 + -0.30)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:33:59 2006