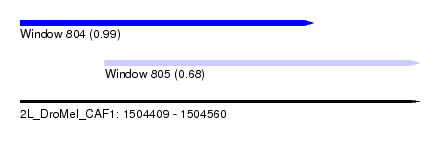
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 1,504,409 – 1,504,560 |
| Length | 151 |
| Max. P | 0.993533 |

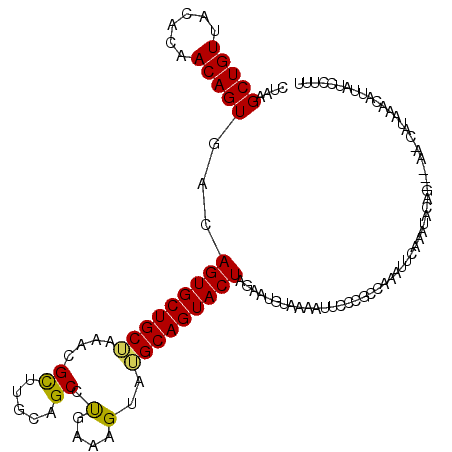
| Location | 1,504,409 – 1,504,520 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 66.73 |
| Mean single sequence MFE | -30.35 |
| Consensus MFE | -12.14 |
| Energy contribution | -12.25 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.41 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.40 |
| SVM decision value | 2.41 |
| SVM RNA-class probability | 0.993533 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1504409 111 + 22407834 --AGAGCUGUUAAGAUAAGAUAACAGUGCUUUUG-------CUGAGCUGUUACACAACAGUGGCAGUGCUGCUAAACGCUUGCAGCCUGAAAGUAUGCAGUACUAGAAUGUAAAAAUUCC --...(((((((........))))))).((..((-------(((.((((((((......))))))))(((((.........)))))...........)))))..)).............. ( -29.40) >DroPse_CAF1 167842 110 + 1 UGUGAUCUGUUAAU-AGACAGAAUCCUGUUAAUGGAACCUAUCGAUAUGUUA-AUUGCAGU-UCAGUGUUGUCAAACCGCAGCAGAGUUC-AGUAGGC---GCGAAAAUGUAGAAAU--- .(((.(((((((((-((........))))))))))).(((((.((..(((..-...)))..-....((((((......))))))....))-.))))))---))..............--- ( -23.40) >DroSec_CAF1 122045 110 + 1 --ACUGCUGUUAAG-UAAGAUAACAGUGCUUAUG-------CUAAGCUGUUACAAAACAGUGACAGUGCUGCUAAACGCUUGCAGCUUGAACGUAAGCAGUACUAGAAUGUAAAAUUCCC --((((((((((..-.....)))))))(((((((-------....((((((((......))))))))(((((.........))))).....))))))))))....((((.....)))).. ( -33.80) >DroSim_CAF1 124521 110 + 1 --ACAGCUGUUAAG-UAAGAUAACAGUGCUUUUG-------CUAAGCUGUUACAAAACAGUGACAGUGCUGCUAAACGCUUGCAGCUUGAACGUAAGCAGUACUAGAAUGUAAAAAUCCC --((((((....))-)........((((((((((-------(...((((((((......))))))))(((((.........)))))......))))).))))))....)))......... ( -29.00) >DroEre_CAF1 128181 99 + 1 -------------A-UAACAUAACAGAGCUGUUG-------CUGAGCUGUAACACAACAGUGACAGUGCUGCAAAACGCUUACAGCCCGAAAGUAUGCAGUACUGUAGUGCACAGUAAGC -------------.-............(((((.(-------(...(((((......))))).(((((((((((....((.....)).(....)..)))))))))))...))))))).... ( -31.70) >DroYak_CAF1 125290 99 + 1 -------------G-UAACGUAACAGUGCUGUAG-------CGUAGCUGUAAAUCAACAGUGACAGUGCUGCAAAAUGUUUACAGCCUGAAAGUAUGCAGUACUGUUAUGCAAAAUACCC -------------.-...((((((((((((((((-------(.(((((((((((...((((......))))......)))))))).)))...)).))))))))))))))).......... ( -34.80) >consensus __A_AGCUGUUAAG_UAACAUAACAGUGCUUUUG_______CUGAGCUGUUACACAACAGUGACAGUGCUGCUAAACGCUUGCAGCCUGAAAGUAUGCAGUACUAGAAUGUAAAAAUCCC ..........................(((................(((((......)))))...(((((((((......................))))))))).....)))........ (-12.14 = -12.25 + 0.11)

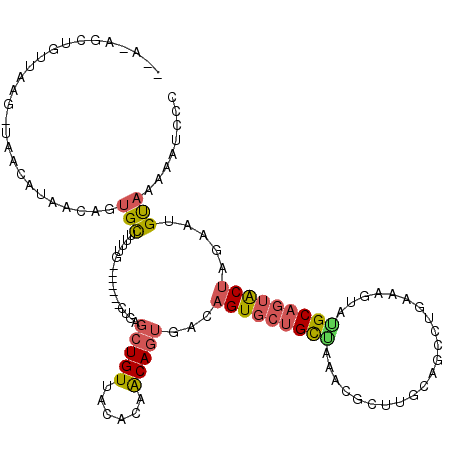
| Location | 1,504,441 – 1,504,560 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 80.57 |
| Mean single sequence MFE | -26.38 |
| Consensus MFE | -15.38 |
| Energy contribution | -15.50 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.678386 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1504441 119 + 22407834 CUGAGCUGUUACACAACAGUGGCAGUGCUGCUAAACGCUUGCAGCCUGAAAGUAUGCAGUACUAGAAUGUAAAAAUUCCGCCAAAUUCAAAUACAGCUGAAGCAUAUGCAUUAUCCUUU .(.((((((..........((((((((((((.....((((.(.....).))))..)))))))).((((......)))).)))).........)))))).).((....)).......... ( -31.31) >DroSec_CAF1 122076 113 + 1 CUAAGCUGUUACAAAACAGUGACAGUGCUGCUAAACGCUUGCAGCUUGAACGUAAGCAGUACUAGAAUGUAAAAUUCCCGCCAAAUUCAAAUACAG------AAUAAACAUUAUCCUUU ....((((((((......))))))))(((((.........)))))(((((.(((.....)))..((((.....))))........)))))......------................. ( -21.10) >DroSim_CAF1 124552 113 + 1 CUAAGCUGUUACAAAACAGUGACAGUGCUGCUAAACGCUUGCAGCUUGAACGUAAGCAGUACUAGAAUGUAAAAAUCCCGCCAAAUUCAAAUACAG------CAUAAACAUUAUCCUUU ....((((((((......))))))))((((......((((((.........)))))).......((((................)))).....)))------)................ ( -23.29) >DroEre_CAF1 128201 119 + 1 CUGAGCUGUAACACAACAGUGACAGUGCUGCAAAACGCUUACAGCCCGAAAGUAUGCAGUACUGUAGUGCACAGUAAGCGCCAAAUUCAAACGUUUUGAAAACAUCAAUAUUAUCCUUU .((((((((......))))).(((((((((((....((.....)).(....)..))))))))))).((((.......))))....(((((.....)))))....)))............ ( -29.60) >DroYak_CAF1 125310 119 + 1 CGUAGCUGUAAAUCAACAGUGACAGUGCUGCAAAAUGUUUACAGCCUGAAAGUAUGCAGUACUGUUAUGCAAAAUACCCGCCAAAUUCGAAUGCAGUGAAACCAUCAAUAUUAUCCUUU ....((((((........((((((((((((((.(...((((.....))))..).))))))))))))))((.........))..........))))))...................... ( -26.60) >consensus CUAAGCUGUUACACAACAGUGACAGUGCUGCUAAACGCUUGCAGCCUGAAAGUAUGCAGUACUAGAAUGUAAAAUUCCCGCCAAAUUCAAAUACAG___AA_CAUAAACAUUAUCCUUU ....(((((......)))))...(((((((((....((((.(.....).)))).)))))))))........................................................ (-15.38 = -15.50 + 0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:37:45 2006