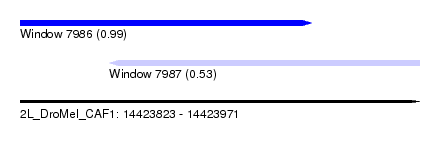
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,423,823 – 14,423,971 |
| Length | 148 |
| Max. P | 0.986290 |

| Location | 14,423,823 – 14,423,931 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 78.32 |
| Mean single sequence MFE | -23.92 |
| Consensus MFE | -8.99 |
| Energy contribution | -9.30 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.72 |
| Structure conservation index | 0.38 |
| SVM decision value | 2.04 |
| SVM RNA-class probability | 0.986290 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
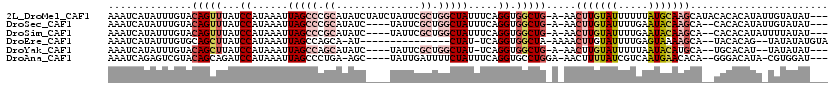
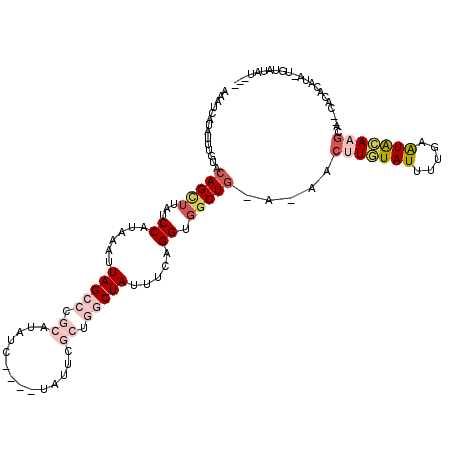
>2L_DroMel_CAF1 14423823 108 + 22407834 AAUCGCCCAUAAAAAAUCGCCAUGUGCACGCAUAUAUACAAUAUGUGUGUAUGCUUGCAUAAAAAAUACAAGUUU-CAGCCACCUGAAAUAGCCAGCGAAUAGAUAGAU ................((((..((((((.((((((((((.....)))))))))).))))))..........((((-(((....))))))).....)))).......... ( -31.20) >DroSec_CAF1 14398 102 + 1 AAUCGUCCAUAAAAAAUCGCCAUGUGCACGCAUAUAUACAAUAUGUGUG--UGCUUGUAUUCAAAAUACAAGUUU-CAGCCACCUGAAAUAGCCAGCGAAUA----GAU ................((((...(..(((((((((.....)))))))))--..)((((((.....))))))((((-(((....))))))).....))))...----... ( -27.70) >DroSim_CAF1 16069 102 + 1 AAUCGUCCAUAAAAAAUCGCCAUGUGCACGCAUAUAUAAAAUAUGUGUG--UGCUUGUAUUCAAAAUACAAGUUU-CAGCCACCUGAAAUAGCCAGCGAAUA----GAU ................((((...(..(((((((((.....)))))))))--..)((((((.....))))))((((-(((....))))))).....))))...----... ( -27.70) >DroYak_CAF1 15787 89 + 1 AAUCGUCCAUAAAAAAUCGCCAU----------AUAUAUA--AUGUGCA--UGCAUGUAUUAAAAAUACAAGUUU-CAGCCACCUGA-AUAGCCAGCGAAUA----GAU ................(((((((----------((.....--)))))..--.((.(((((.....)))))...((-(((....))))-)..))..))))...----... ( -12.80) >DroAna_CAF1 6632 102 + 1 AGCCGUCCAUAAAAAAGCGCUAGAUGUGCGCGUAUCCACG-UAUGUCCC--UGUGUUCAUUGACGAUAAAAGUUUCCAGGCACCUGAAAUAGAAAAUCAAUA----GCU (((((((.((.....(((((..((((((((.(....).))-))))))..--.))))).)).))))......(((((.((....)))))))............----))) ( -20.20) >consensus AAUCGUCCAUAAAAAAUCGCCAUGUGCACGCAUAUAUACAAUAUGUGUG__UGCUUGUAUUAAAAAUACAAGUUU_CAGCCACCUGAAAUAGCCAGCGAAUA____GAU ................((((.(((((....)))))........((.((....((((((((.....)))))))).....)))).............)))).......... ( -8.99 = -9.30 + 0.31)

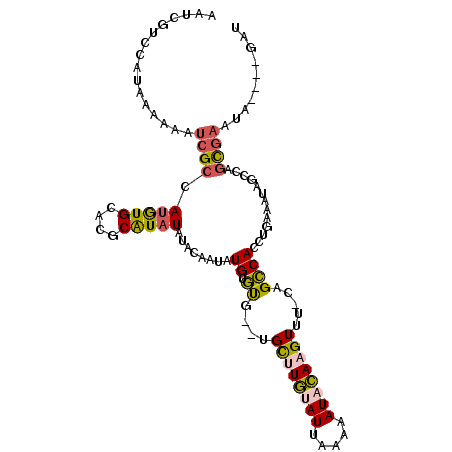
| Location | 14,423,856 – 14,423,971 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.86 |
| Mean single sequence MFE | -23.32 |
| Consensus MFE | -10.59 |
| Energy contribution | -12.34 |
| Covariance contribution | 1.75 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.45 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.526360 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14423856 115 - 22407834 AAAUCAUAUUUGUACAGUUUAUCCAUAAAUUAGCCCGCAUAUCUAUCUAUUCGCUGGCUAUUUCAGGUGGCUG-A-AACUUGUAUUUUUUAUGCAAGCAUACACACAUAUUGUAUAU--- ..........(((((((((((.((((....(((((.((..............)).)))))......)))).))-)-).(((((((.....)))))))............))))))).--- ( -24.54) >DroSec_CAF1 14431 109 - 1 AAAUCAUAUUUGUACAGUUUAUCCAUAAAUUAGCCCGCAUAUC----UAUUCGCUGGCUAUUUCAGGUGGCUG-A-AACUUGUAUUUUGAAUACAAGCA--CACACAUAUUGUAUAU--- ..........(((((((((((.((((....(((((.((.....----.....)).)))))......)))).))-)-).(((((((.....)))))))..--........))))))).--- ( -24.40) >DroSim_CAF1 16102 109 - 1 AAAUCAUAUUUGUACAGUUUAUCCAUAAAUUAGCCCGCAUAUC----UAUUCGCUGGCUAUUUCAGGUGGCUG-A-AACUUGUAUUUUGAAUACAAGCA--CACACAUAUUUUAUAU--- .....((((.(((.(((((...((......(((((.((.....----.....)).))))).....)).)))))-.-..(((((((.....)))))))..--.))).)))).......--- ( -22.30) >DroEre_CAF1 15746 98 - 1 AAAUCAUAUUUGUGCAGCUUAUCCAUAAAUUAGCCAGCA-AU---------------CUAU-UCAGGUGGCUA-AAAACUUGUAUUUUGAGUAAAAGCA--UACACAG--UAUAUAUGUA ....((((((((((..((((.........(((((((.(.-..---------------....-...).))))))-)..(((((.....)))))..)))).--..)))))--...))))).. ( -20.10) >DroYak_CAF1 15810 106 - 1 AAAUCAUAUUUGUACAGCUUAUCCAUAAAUUAGCCAGCAUAUC----UAUUCGCUGGCUAU-UCAGGUGGCUG-A-AACUUGUAUUUUUAAUACAUGCA--UGCACAU--UAUAUAU--- .....((((.(((.(((((((.((((....((((((((.....----.....)))))))).-....)))).))-)-....(((((.....))))).)).--)).))).--.))))..--- ( -25.20) >DroAna_CAF1 6665 108 - 1 AAAUCAGAGUCGUACAGCAGAUCCAUAAAUUAGCCCUGA-AGC----UAUUGAUUUUCUAUUUCAGGUGCCUGGA-AACUUUUAUCGUCAAUGAACACA--GGGACAUA-CGUGGAU--- ....................((((((.......(((((.-...----(((((((......((((((....)))))-).........)))))))....))--))).....-.))))))--- ( -23.38) >consensus AAAUCAUAUUUGUACAGCUUAUCCAUAAAUUAGCCCGCAUAUC____UAUUCGCUGGCUAUUUCAGGUGGCUG_A_AACUUGUAUUUUGAAUACAAGCA__CACACAUA_UGUAUAU___ ..............(((((...((......(((((.((..............)).))))).....)).))))).....(((((((.....)))))))....................... (-10.59 = -12.34 + 1.75)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:33:56 2006