| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,422,254 – 14,422,356 |
| Length | 102 |
| Max. P | 0.927599 |

| Location | 14,422,254 – 14,422,356 |
|---|---|
| Length | 102 |
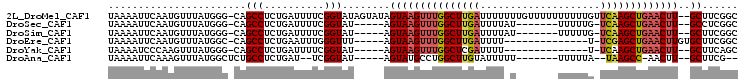
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 79.97 |
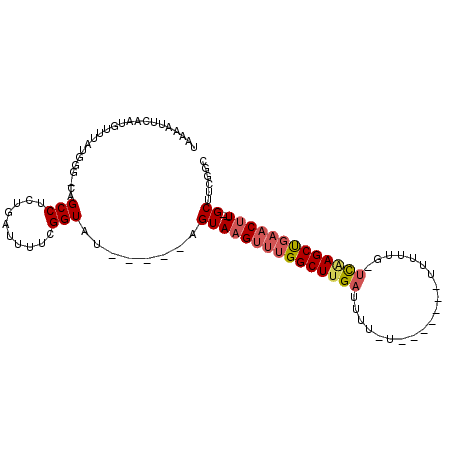
| Mean single sequence MFE | -24.34 |
| Consensus MFE | -11.49 |
| Energy contribution | -12.10 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.17 |
| Mean z-score | -3.05 |
| Structure conservation index | 0.47 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.927599 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14422254 102 - 22407834 UAAAAUUCAAUGUUUAUGGG-CAGCCUCUGAUUUUCGGUAUAGUAUAGUAAGUUUGGCUUGAUUUUUUUGUUUUUUUUUUGUUCAAGCUGAACUU--GCUUCGGC ..........((((....))-))(((.(((.....)))....(...((((((((..((((((....................))))))..)))))--))).)))) ( -25.05) >DroSec_CAF1 12847 89 - 1 UAAAAUUCAAUGUUUAUGGG-CAGCCUCUGAUUUUCGGUAU-----AGUAAGUUUGGCUUGAUUUUAU-------UUUUUG-UCAAGCUGAACUU--GCCUCGGC ..........((((....))-))(((.(((.....)))...-----.(((((((..(((((((.....-------.....)-))))))..)))))--))...))) ( -25.90) >DroSim_CAF1 14535 89 - 1 UAAAAUUCAAUGUUUAUGGG-CAGCCUCUGAUUUUCGGUAU-----AGUAAGUUUGGCUUGAUUUUAU-------UUUUUG-UCAAGCUGAACUU--GCUUCGGC ..........((((....))-))(((.(((.....)))...-----((((((((..(((((((.....-------.....)-))))))..)))))--)))..))) ( -26.80) >DroEre_CAF1 14159 84 - 1 UAAAAUUCAAUGUUUAUGGC-CAGCCUCUGAAUUUGGGUUU-----AGUAAGUUUGGCUUGAUUUU--------------U-UCGAGCUGAACUUGUGCUUCGGC ..((((((((.(((((.((.-...))..)))))))))))))-----..((((((..((((((....--------------.-))))))..)))))).((....)) ( -27.30) >DroYak_CAF1 14249 82 - 1 UAAAAUCCCAAGUUUAUGGG-CAGCCUCUGAUUUUCGGUAU-----AGUAAGUUUGGCUCGAUUUU--------------U-UCAAGCUGAACUU--GCUUCAGC ......((((......))))-......((((..........-----((((((((..(((.((....--------------.-)).)))..)))))--))))))). ( -22.80) >DroAna_CAF1 5215 84 - 1 UAAAAUUCAAAGUUUAUGGCUCUGCCUCUGAU--UCGGUAU-----AGUAUGCCUGGCUUGUAUUUUU-------UUUUUA--UAAGCC-AACUU--GCUUCG-- .........(((((...(((.(((((......--..)))..-----))...))).((((((((.....-------....))--))))))-)))))--......-- ( -18.20) >consensus UAAAAUUCAAUGUUUAUGGG_CAGCCUCUGAUUUUCGGUAU_____AGUAAGUUUGGCUUGAUUUU_U_______UUUUUG_UCAAGCUGAACUU__GCUUCGGC .......................(((..........)))........(((((((((((((((....................)))))))))))))..))...... (-11.49 = -12.10 + 0.61)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:33:54 2006