| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,413,888 – 14,413,987 |
| Length | 99 |
| Max. P | 0.521942 |

| Location | 14,413,888 – 14,413,987 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.61 |
| Mean single sequence MFE | -23.63 |
| Consensus MFE | -14.28 |
| Energy contribution | -15.28 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.60 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.521942 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
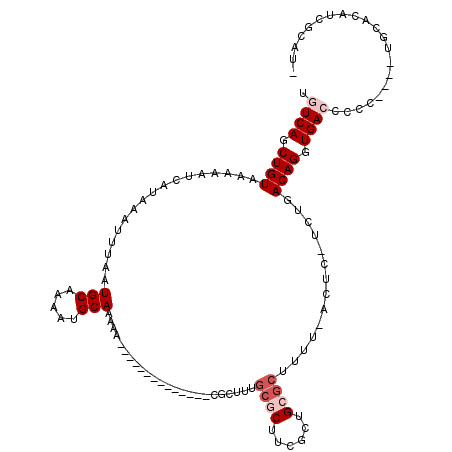
>2L_DroMel_CAF1 14413888 99 + 22407834 UGUCAGCUGUAAAAAUCAUAAAUUUAAUGCAAAAUGCAAAAA--------------CGCUUUGCGCUCCGCUGGGCUUUU-GCUC-UCUGACAGGUGACCCCU----UGCACCUCGCAG- ((((((..((((((.............(((.....)))....--------------.(((..(((...)))..)))))))-))..-.))))))((((......----..))))......- ( -25.50) >DroVir_CAF1 18144 95 + 1 UGUCAGCUGUAAAAAUCAUAAAUUUAAUGCAAAAUGCAAAAA--------------CGU-UUGCGCUUCGUUGCGCUUUU-ACUC-U-UGACAGGUGACCCUG------CUCAUCGCCC- ((((((..((((((.............(((.....)))....--------------...-..((((......))))))))-))..-)-)))))(((((.....------....))))).- ( -24.40) >DroPse_CAF1 365 104 + 1 UGUCAGCUGUAAAAAUCAUAAAUUUAAUGCAAAAUGCAAAAA--------------CGCUUUGCGCUUCGCUGCGCUUUU-ACUCCACUGACAGGUGAACCCUCUCUCGCACCUCUCCA- ((((((..((((((.............(((.....)))....--------------......((((......))))))))-))....))))))((((............))))......- ( -23.80) >DroEre_CAF1 5529 99 + 1 UGUCAGCUGUAAAAAUCAUAAAUUUAAUGCAAAAUGCAAAAA--------------CGCUUUGCGCUUCGCUGGGCUUUUGGCUC-CCUGACAGGUGA-CCCC----UGCACCUCGCAU- .((((.((((..................((.((.((((((..--------------...)))))).)).)).((((.....))))-....)))).)))-)...----(((.....))).- ( -22.80) >DroWil_CAF1 88966 115 + 1 UGUCAGCUGUAAAAAUCAUAAAUUUAAUGCAAAAUGCAAAAAAAAGAAAGAAGGAGCUCGUUGAGCUUCGUUGCGCUUUU-ACUC-GCUGACAGGUGACCCUCCA--UUCAUAUAGCAU- (((((((.((((((.............(((.....)))..............(((((((...))))))).......))))-))..-))))))).((((.......--.)))).......- ( -27.30) >DroMoj_CAF1 38004 95 + 1 UGUCAGCUGUAAAAAUCAUAAAUUUAAUGCAAAAUGCAAAAA--------------CGU-UU-CGCUUCGUUGCGCUUUU-ACUC-U-UGACAGGUGACCCCG------CUCAGCCCCUC ((((((..((((((.............(((.....)))....--------------...-..-(((......))).))))-))..-)-)))))(((((.....------.))).)).... ( -18.00) >consensus UGUCAGCUGUAAAAAUCAUAAAUUUAAUGCAAAAUGCAAAAA______________CGCUUUGCGCUUCGCUGCGCUUUU_ACUC_UCUGACAGGUGACCCCC____UGCACAUCGCAU_ .((((.((((.................(((.....)))........................((((......))))..............)))).))))..................... (-14.28 = -15.28 + 1.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:33:52 2006