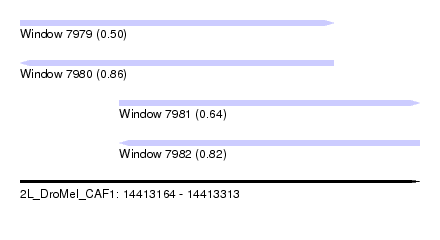
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,413,164 – 14,413,313 |
| Length | 149 |
| Max. P | 0.864296 |

| Location | 14,413,164 – 14,413,281 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.76 |
| Mean single sequence MFE | -31.07 |
| Consensus MFE | -22.77 |
| Energy contribution | -22.80 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.73 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14413164 117 + 22407834 CCGCUGCUGUUGGUGC---UCUCUUCAUCUUUUCUGCUGUCGCCUGUCGCCGCCUCUCAUGCGGCAACAGCAACUGUCGCCGUUGCUGUCACUUGAUGUCGCAACGCAUUAGAAUAUGUU ....((((((((((((---...................).))))....(((((.......)))))))))))).(((..((.(((((.(((....)))...)))))))..)))........ ( -31.91) >DroPse_CAF1 28 89 + 1 -------------------------------GUCGCCUGUCGCCUGUCGCCGCCUCUCAUGCGGCAACAGCAACUGUCGCCGUUGCUGUCACUUGAUGUCGCAGCGCAUUAGAAUAUGUU -------------------------------...((((((.((..((((((((.......))))).((((((((.......)))))))).....))))).)))).))............. ( -29.80) >DroSec_CAF1 3731 120 + 1 CCGCUGCUGCUGGUGCUCCUCUCUUCAUCUUUUCUGCUGUCGCCUGUCGCCGCCUCUCAUGCGGCAACAGCAACUGUCGCCGUUGCUGUCACUUGAUGUCGCAACGCAUUAGAAUAUGUU ..((.((.((.(((((..(................)..).)))).)).)).)).(((.(((((((.((((...)))).)))(((((.(((....)))...))))))))).)))....... ( -32.69) >DroEre_CAF1 4908 113 + 1 ----UGCCGCUGGUGC---UCUCUUCAUCUUUGCUGCUGUCGCCUGUCGCCGCCUCUCAUGCGGCAACAGCAACUGUCGCCGUUGCUGUCACUUGAUGUCGCAACGCAUUAGAAUAUGUU ----.....(((((((---.............((.((.((.((.(((.(((((.......))))).))))).)).)).)).(((((.(((....)))...))))))))))))........ ( -34.50) >DroWil_CAF1 88531 91 + 1 -----------------------------UUUUACCCUCCACCCUGUCACCGCCUCUCAUGCGGCAACAGCAACUGUCGCCGUUGCUGUCAGUUCAUGUCGCAACGCAUUAGAAUAUGUU -----------------------------.........................(((.(((((((.((((...)))).)))(((((.(.((.....)).)))))))))).)))....... ( -23.00) >DroYak_CAF1 5014 114 + 1 C---UGCUGCUGGUGC---UCUCUUCAUCUUUGCUGCUGUCGCCUGUCGCCGCCUCUCAUGCGGCAACAGCAACUGUCGCCGUUGCUGUCACUUGAUGUCGCAACGCAUUAGAAUAUGUU .---.....(((((((---.............((.((.((.((.(((.(((((.......))))).))))).)).)).)).(((((.(((....)))...))))))))))))........ ( -34.50) >consensus ____UGCUGCUGGUGC___UCUCUUCAUCUUUUCUGCUGUCGCCUGUCGCCGCCUCUCAUGCGGCAACAGCAACUGUCGCCGUUGCUGUCACUUGAUGUCGCAACGCAUUAGAAUAUGUU ...........................................((((.(((((.......))))).))))...(((..((.(((((.(.((.....)).))))))))..)))........ (-22.77 = -22.80 + 0.03)



| Location | 14,413,164 – 14,413,281 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.76 |
| Mean single sequence MFE | -36.17 |
| Consensus MFE | -29.33 |
| Energy contribution | -29.72 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.864296 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14413164 117 - 22407834 AACAUAUUCUAAUGCGUUGCGACAUCAAGUGACAGCAACGGCGACAGUUGCUGUUGCCGCAUGAGAGGCGGCGACAGGCGACAGCAGAAAAGAUGAAGAGA---GCACCAACAGCAGCGG ..............((((((..((((..((((((((((((....).)))))))))(((((.(....)))))).))..((....))......))))....(.---....)....)))))). ( -37.70) >DroPse_CAF1 28 89 - 1 AACAUAUUCUAAUGCGCUGCGACAUCAAGUGACAGCAACGGCGACAGUUGCUGUUGCCGCAUGAGAGGCGGCGACAGGCGACAGGCGAC------------------------------- ............(((((((..((.....))..))))..........((((((((((((((.(....)))))))))).)))))..)))..------------------------------- ( -33.00) >DroSec_CAF1 3731 120 - 1 AACAUAUUCUAAUGCGUUGCGACAUCAAGUGACAGCAACGGCGACAGUUGCUGUUGCCGCAUGAGAGGCGGCGACAGGCGACAGCAGAAAAGAUGAAGAGAGGAGCACCAGCAGCAGCGG ..............((((((..((((..((((((((((((....).)))))))))(((((.(....)))))).))..((....))......)))).........((....)).)))))). ( -41.10) >DroEre_CAF1 4908 113 - 1 AACAUAUUCUAAUGCGUUGCGACAUCAAGUGACAGCAACGGCGACAGUUGCUGUUGCCGCAUGAGAGGCGGCGACAGGCGACAGCAGCAAAGAUGAAGAGA---GCACCAGCGGCA---- ......((((....((((((..(((...)))...))))))((..(.((((((((((((((.(....)))))))))).))))).)..))........)))).---((....))....---- ( -37.90) >DroWil_CAF1 88531 91 - 1 AACAUAUUCUAAUGCGUUGCGACAUGAACUGACAGCAACGGCGACAGUUGCUGUUGCCGCAUGAGAGGCGGUGACAGGGUGGAGGGUAAAA----------------------------- ..(((.((((.(((((((((..((.....))...)))))((((((((...)))))))))))).))))..(....)...)))..........----------------------------- ( -29.40) >DroYak_CAF1 5014 114 - 1 AACAUAUUCUAAUGCGUUGCGACAUCAAGUGACAGCAACGGCGACAGUUGCUGUUGCCGCAUGAGAGGCGGCGACAGGCGACAGCAGCAAAGAUGAAGAGA---GCACCAGCAGCA---G ......((((....((((((..(((...)))...))))))((..(.((((((((((((((.(....)))))))))).))))).)..))........)))).---((....))....---. ( -37.90) >consensus AACAUAUUCUAAUGCGUUGCGACAUCAAGUGACAGCAACGGCGACAGUUGCUGUUGCCGCAUGAGAGGCGGCGACAGGCGACAGCAGAAAAGAUGAAGAGA___GCACCAGCAGCA____ ............((((((((..((.....))...))))).)))...((((((((((((((.(....)))))))))).)))))...................................... (-29.33 = -29.72 + 0.39)

| Location | 14,413,201 – 14,413,313 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.10 |
| Mean single sequence MFE | -32.13 |
| Consensus MFE | -21.95 |
| Energy contribution | -22.23 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.16 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.639180 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
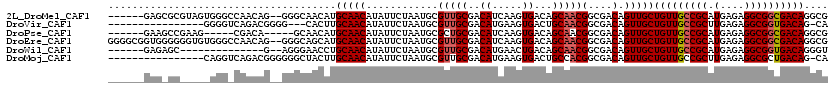
>2L_DroMel_CAF1 14413201 112 + 22407834 CGCCUGUCGCCGCCUCUCAUGCGGCAACAGCAACUGUCGCCGUUGCUGUCACUUGAUGUCGCAACGCAUUAGAAUAUGUUGCAUGUUGCCC--CUGUUGGCCCACUACGCGCUC------ .((.(((.(..(((...((.(.((((((((((((..((..((((((.(((....)))...)))))).....))....))))).))))))))--.))..)))...).))).))..------ ( -36.70) >DroVir_CAF1 17660 100 + 1 UG-CUGUCACCGCCUCUCAAGCGGCAACAGCAACUGUCGCCGUUGCAGUCACUUCAUGUCGCAACGCAUUAGAAUAUGUUGCAAGUG---CCCCGUCUGACCCC---------------- ((-((((..((((.......))))..))))))...(((((.(((((.(.((.....)).))))))))...(((.......((....)---)....))))))...---------------- ( -26.80) >DroPse_CAF1 37 104 + 1 CGCCUGUCGCCGCCUCUCAUGCGGCAACAGCAACUGUCGCCGUUGCUGUCACUUGAUGUCGCAGCGCAUUAGAAUAUGUUGCAUGUUGC-----UGUCG-----CUUCGGCUUC------ .(((....(((((.......))))).(((((((((((.......(((((.((.....)).)))))((((......)))).))).)))))-----)))..-----....)))...------ ( -37.20) >DroEre_CAF1 4941 118 + 1 CGCCUGUCGCCGCCUCUCAUGCGGCAACAGCAACUGUCGCCGUUGCUGUCACUUGAUGUCGCAACGCAUUAGAAUAUGUUGCAUGCUGCCC--CUGUUGGCCCACACCCCCCACCGCCCC .(((.((((((((.......))))).((((((((.......)))))))).....)))...((((((.((....)).)))))).........--.....)))................... ( -31.60) >DroWil_CAF1 88542 98 + 1 ACCCUGUCACCGCCUCUCAUGCGGCAACAGCAACUGUCGCCGUUGCUGUCAGUUCAUGUCGCAACGCAUUAGAAUAUGUUGCAGGUUCCCU--C--------------GCUCUC------ ..(((((.((....(((.(((((((.((((...)))).)))(((((.(.((.....)).)))))))))).)))....)).)))))......--.--------------......------ ( -31.00) >DroMoj_CAF1 37548 103 + 1 UG-CUGUCAGCGCCUCUCAAGCGGCAACAGCAACUGUCGCCGUGGCAGUCACUUCAUGUCGCAACGCAUUAGAAUAUGUUGCAAGUAGCCCCCCGUCUGACCUG---------------- ..-..(((((((........(((((.((((...)))).)))))(((.(.((.....)).)((((((.((....)).)))))).....)))...)).)))))...---------------- ( -29.50) >consensus CGCCUGUCACCGCCUCUCAUGCGGCAACAGCAACUGUCGCCGUUGCUGUCACUUCAUGUCGCAACGCAUUAGAAUAUGUUGCAUGUUGCCC__CUGUUG_CCC_____GC_CUC______ ....(((.((....(((.(((((((.((((...)))).)))(((((.(.((.....)).)))))))))).)))....)).)))..................................... (-21.95 = -22.23 + 0.28)


| Location | 14,413,201 – 14,413,313 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.10 |
| Mean single sequence MFE | -38.75 |
| Consensus MFE | -25.85 |
| Energy contribution | -26.10 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.821929 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14413201 112 - 22407834 ------GAGCGCGUAGUGGGCCAACAG--GGGCAACAUGCAACAUAUUCUAAUGCGUUGCGACAUCAAGUGACAGCAACGGCGACAGUUGCUGUUGCCGCAUGAGAGGCGGCGACAGGCG ------..((((((.((..(((.....--.))).))))))......((((.(((((((((..(((...)))...)))))((((((((...)))))))))))).)))))).((.....)). ( -40.50) >DroVir_CAF1 17660 100 - 1 ----------------GGGGUCAGACGGGG---CACUUGCAACAUAUUCUAAUGCGUUGCGACAUGAAGUGACUGCAACGGCGACAGUUGCUGUUGCCGCUUGAGAGGCGGUGACAG-CA ----------------...(((...((..(---((.(((((((.(((....))).)))))))(((...)))..)))..))..)))...((((((..(((((.....)))))..))))-)) ( -37.80) >DroPse_CAF1 37 104 - 1 ------GAAGCCGAAG-----CGACA-----GCAACAUGCAACAUAUUCUAAUGCGCUGCGACAUCAAGUGACAGCAACGGCGACAGUUGCUGUUGCCGCAUGAGAGGCGGCGACAGGCG ------...(((((.(-----((.((-----((..(((.............))).)))))).).))..((((((((((((....).)))))))))(((((.(....)))))).)).))). ( -37.82) >DroEre_CAF1 4941 118 - 1 GGGGCGGUGGGGGGUGUGGGCCAACAG--GGGCAGCAUGCAACAUAUUCUAAUGCGUUGCGACAUCAAGUGACAGCAACGGCGACAGUUGCUGUUGCCGCAUGAGAGGCGGCGACAGGCG .(((..(((..(.((((..(((.....--.))).)))).)..)))..)))...((((..(........)..))(((((((....).))))))((((((((.(....)))))))))..)). ( -45.10) >DroWil_CAF1 88542 98 - 1 ------GAGAGC--------------G--AGGGAACCUGCAACAUAUUCUAAUGCGUUGCGACAUGAACUGACAGCAACGGCGACAGUUGCUGUUGCCGCAUGAGAGGCGGUGACAGGGU ------......--------------.--......((((..((...((((.(((((((((..((.....))...)))))((((((((...)))))))))))).))))...))..)))).. ( -35.90) >DroMoj_CAF1 37548 103 - 1 ----------------CAGGUCAGACGGGGGGCUACUUGCAACAUAUUCUAAUGCGUUGCGACAUGAAGUGACUGCCACGGCGACAGUUGCUGUUGCCGCUUGAGAGGCGCUGACAG-CA ----------------...(((((.((((.((.((((((((((.(((....))).)))))......))))).)).)).(((((((((...))))))))).........)))))))..-.. ( -35.40) >consensus ______GAG_GC_____GGG_CAACAG__GGGCAACAUGCAACAUAUUCUAAUGCGUUGCGACAUCAAGUGACAGCAACGGCGACAGUUGCUGUUGCCGCAUGAGAGGCGGCGACAGGCG ......................................(((((............(((((..((.....))...)))))(....).)))))(((((((((.(....)))))))))).... (-25.85 = -26.10 + 0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:33:51 2006