| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,411,530 – 14,411,641 |
| Length | 111 |
| Max. P | 0.861162 |

| Location | 14,411,530 – 14,411,624 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 91.70 |
| Mean single sequence MFE | -18.82 |
| Consensus MFE | -13.89 |
| Energy contribution | -14.22 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.861162 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14411530 94 + 22407834 AUUAACCACACAAAAAGUAUAUGUCGUUUUCGCUCUCUGGGUAAACCACAAAAUUAAGUGGCUAACAGUAUUUUGUGGCCACAACACAUUACAA ................(((.((((.(((.........(((.....))).........(((((((.(((....)))))))))))))))))))).. ( -19.50) >DroSec_CAF1 2089 94 + 1 AUUAACCACACAAAAAGUAUAUGUCGUUCUCGCUCUCCGGGUAAACCACAAAAUUAAGUGGCUAACAGUAUUUUGUGGCCACAACACAUUACAA ................(((.((((.(((..........((.....))..........(((((((.(((....)))))))))))))))))))).. ( -19.30) >DroSim_CAF1 3169 94 + 1 AUUAACCACACAAAAAGUAUAUGUCGUUCUCGCUCUCCGGGUAAACCACAAAAUUAAGUGGCUAACAGUAUUUUGUGGCCACAACACAUUACAA ................(((.((((.(((..........((.....))..........(((((((.(((....)))))))))))))))))))).. ( -19.30) >DroEre_CAF1 3324 93 + 1 AUUAACCACACAA-AAGUAUAUGUCGUUCUCGCUCUCUGGGCAAACCACAAAAUUAAGUGGCUAACAGUAUUUUGUGGCCACAACACAUUACAA .............-..(((.((((.(((...(((.....)))...............(((((((.(((....)))))))))))))))))))).. ( -20.80) >DroYak_CAF1 3405 84 + 1 AUUAACCACACAAAAAGUAUAUAUCGCUCCCGCUCUCUGGGUGAACCGCAAAAUUAAG----------UAUUUUGUGGCCACAACACAUUACAA ................(((....((((((.........)))))).(((((((((....----------.)))))))))...........))).. ( -15.20) >consensus AUUAACCACACAAAAAGUAUAUGUCGUUCUCGCUCUCUGGGUAAACCACAAAAUUAAGUGGCUAACAGUAUUUUGUGGCCACAACACAUUACAA ................(((.((((.(((...(((.....)))...(((((((((...((.....))...)))))))))....)))))))))).. (-13.89 = -14.22 + 0.33)



| Location | 14,411,549 – 14,411,641 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 78.35 |
| Mean single sequence MFE | -21.03 |
| Consensus MFE | -11.38 |
| Energy contribution | -12.70 |
| Covariance contribution | 1.32 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.792177 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
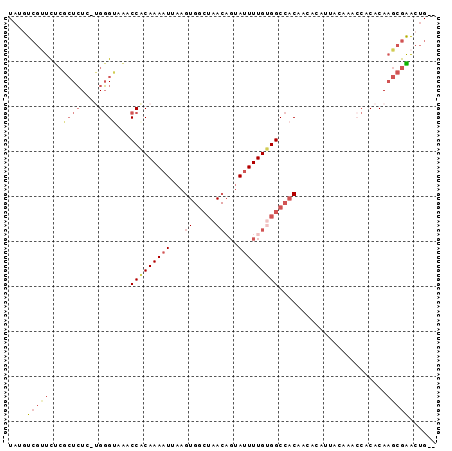
>2L_DroMel_CAF1 14411549 92 + 22407834 UAUGUCGUUUUCGCUCUC-UGGGUAAACCACAAAAUUAAGUGGCUAACAGUAUUUUGUGGCCACAACACAUUACAAACCACACAAGCGAACUG-- .........((((((...-(((.....))).........(((((((.(((....))))))))))....................))))))...-- ( -21.60) >DroVir_CAF1 14453 73 + 1 UAUAU---UUU-UGUGUGUUGUGCCGACCACAAAAUUAAGUGGCUAACUGAA-UUUGCGGCCACA---------------CACAUGUGGGCAG-- .....---...-(((((((.(.((((......(((((.(((.....))).))-))).))))))))---------------)))).........-- ( -19.10) >DroSec_CAF1 2108 92 + 1 UAUGUCGUUCUCGCUCUC-CGGGUAAACCACAAAAUUAAGUGGCUAACAGUAUUUUGUGGCCACAACACAUUACAAACCACACAAGCGAACUG-- ..........(((((...-.((.....))..........(((((((.(((....))))))))))....................)))))....-- ( -20.70) >DroSim_CAF1 3188 92 + 1 UAUGUCGUUCUCGCUCUC-CGGGUAAACCACAAAAUUAAGUGGCUAACAGUAUUUUGUGGCCACAACACAUUACAAACCACACAAGCGAACUG-- ..........(((((...-.((.....))..........(((((((.(((....))))))))))....................)))))....-- ( -20.70) >DroEre_CAF1 3342 94 + 1 UAUGUCGUUCUCGCUCUC-UGGGCAAACCACAAAAUUAAGUGGCUAACAGUAUUUUGUGGCCACAACACAUUACAAACCCCACAAGCGAACUGUU ..........(((((...-((((....((((........))))......(((...((((.......)))).)))....))))..)))))...... ( -21.80) >DroYak_CAF1 3424 82 + 1 UAUAUCGCUCCCGCUCUC-UGGGUGAACCGCAAAAUUAAG----------UAUUUUGUGGCCACAACACAUUACAAAACCCACAAGCGGACUG-- ..........(((((...-(((((...(((((((((....----------.))))))))).................)))))..)))))....-- ( -22.25) >consensus UAUGUCGUUCUCGCUCUC_UGGGUAAACCACAAAAUUAAGUGGCUAACAGUAUUUUGUGGCCACAACACAUUACAAACCACACAAGCGAACUG__ ....(((((...((((....))))...(((((((((...((.....))...)))))))))........................)))))...... (-11.38 = -12.70 + 1.32)


| Location | 14,411,549 – 14,411,641 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 78.35 |
| Mean single sequence MFE | -25.62 |
| Consensus MFE | -14.81 |
| Energy contribution | -16.28 |
| Covariance contribution | 1.46 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.843163 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
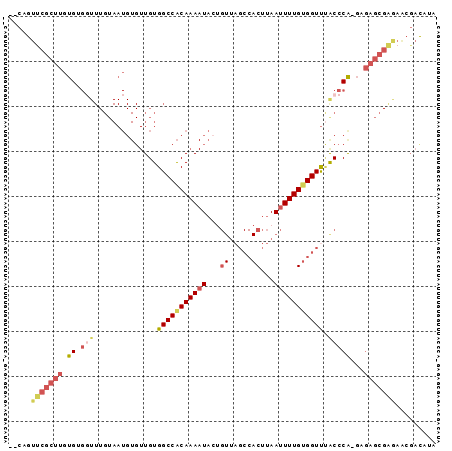
>2L_DroMel_CAF1 14411549 92 - 22407834 --CAGUUCGCUUGUGUGGUUUGUAAUGUGUUGUGGCCACAAAAUACUGUUAGCCACUUAAUUUUGUGGUUUACCCA-GAGAGCGAAAACGACAUA --...(((((((.(((((.(..(((....)))..)))))).....(((..((((((........))))))....))-).)))))))......... ( -27.10) >DroVir_CAF1 14453 73 - 1 --CUGCCCACAUGUG---------------UGUGGCCGCAAA-UUCAGUUAGCCACUUAAUUUUGUGGUCGGCACAACACACA-AAA---AUAUA --.........((((---------------(((.((((....-........(((((........)))))))))...)))))))-...---..... ( -22.30) >DroSec_CAF1 2108 92 - 1 --CAGUUCGCUUGUGUGGUUUGUAAUGUGUUGUGGCCACAAAAUACUGUUAGCCACUUAAUUUUGUGGUUUACCCG-GAGAGCGAGAACGACAUA --...(((((((.(((((.(..(((....)))..)))))).....(((..((((((........))))))....))-).)))))))......... ( -26.10) >DroSim_CAF1 3188 92 - 1 --CAGUUCGCUUGUGUGGUUUGUAAUGUGUUGUGGCCACAAAAUACUGUUAGCCACUUAAUUUUGUGGUUUACCCG-GAGAGCGAGAACGACAUA --...(((((((.(((((.(..(((....)))..)))))).....(((..((((((........))))))....))-).)))))))......... ( -26.10) >DroEre_CAF1 3342 94 - 1 AACAGUUCGCUUGUGGGGUUUGUAAUGUGUUGUGGCCACAAAAUACUGUUAGCCACUUAAUUUUGUGGUUUGCCCA-GAGAGCGAGAACGACAUA .....(((((((...(((((..(((.((...(((((.(((......)))..)))))...)).)))..)...)))).-..)))))))......... ( -27.40) >DroYak_CAF1 3424 82 - 1 --CAGUCCGCUUGUGGGUUUUGUAAUGUGUUGUGGCCACAAAAUA----------CUUAAUUUUGCGGUUCACCCA-GAGAGCGGGAGCGAUAUA --...(((((((.(((((.((((((.(((((.((....)).))))----------)......))))))...)))))-..)))))))......... ( -24.70) >consensus __CAGUUCGCUUGUGUGGUUUGUAAUGUGUUGUGGCCACAAAAUACUGUUAGCCACUUAAUUUUGUGGUUUACCCA_GAGAGCGAGAACGACAUA .....(((((((.((.(((..............(((((((((((...((.....))...))))))))))).)))))...)))))))......... (-14.81 = -16.28 + 1.46)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:33:47 2006