
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,403,453 – 14,403,678 |
| Length | 225 |
| Max. P | 0.965002 |

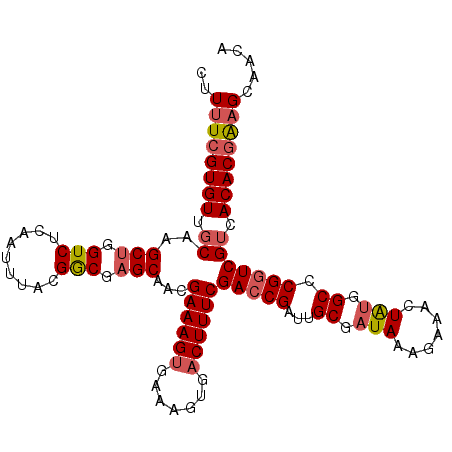
| Location | 14,403,453 – 14,403,562 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 93.76 |
| Mean single sequence MFE | -34.80 |
| Consensus MFE | -28.68 |
| Energy contribution | -29.44 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.936017 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14403453 109 - 22407834 CUUUUCGUGUUGCAAGCUGGUCUCAAUUUACGGCGAGCACCGAAAGUGAAAGUGACUUUCGACCGAUUGCGAUAAAGAAACUAUGGCCCGUUCGUCACACGAAGCAACA ..((((((((.((.(((.((((....((((..((((....(((((((.......))))))).....))))..))))........)))).))).)).))))))))..... ( -30.00) >DroSec_CAF1 24557 109 - 1 CUUUUCGUGUUGCAAGCUGGUCUCAAUUUACGGCGAGCAACGAAAGUGAAAGUGACUUUCGACCGAUUGCGAUAAAGAAACUGAGGCCCGGUCGUCACACGAAGCAACA ..((((((((.((.(.(.(((((((.((((..((((....(((((((.......))))))).....))))..)))).....))))))).).).)).))))))))..... ( -33.80) >DroSim_CAF1 23456 109 - 1 CUUUUCGUGUGGCAAGCUGGUCUCAAUUUACGGCGAGCAACGAAAGUGAAAGUGACUUUCGACCGAUUGCGAUAAAGAAACUGUGGCCCGGUCGUCACACGAAGCAACA ..(((((((((((..(((.(((.........))).)))...((((((.......))))))(((((...((.(((.......))).)).))))))))))))))))..... ( -39.80) >DroEre_CAF1 24658 109 - 1 CUUUUCGUGUUGCAAGCCGGUCUCAAUUCACGACGAGCACCGAAAGUGAAAGUGCCUUUCGACCGAUUGCGAUAAAGAAAUUAUGGCCCGGUCGGCACACGGAGCAACA .......((((((...(((.......(((((..((.....))...))))).(((((....(((((...((.((((.....)))).)).)))))))))).))).)))))) ( -36.30) >DroYak_CAF1 24635 109 - 1 CUUUUCGUGUUGCAAGCUGGUCUCAAUUUACGGCGAGCAACGAAAGUGAAAGUGACUUUCGACCGAUUGCGAUACAGAAAUUAUGGCCCGGUCGUCACACUGCGCAACA .......((((((..((((...........))))..(((((....))....(((((....(((((...((.(((.(....)))).)).))))))))))..))))))))) ( -34.10) >consensus CUUUUCGUGUUGCAAGCUGGUCUCAAUUUACGGCGAGCAACGAAAGUGAAAGUGACUUUCGACCGAUUGCGAUAAAGAAACUAUGGCCCGGUCGUCACACGAAGCAACA ..((((((((.((..(((.(((.........))).)))...((((((.......))))))(((((...((.(((.......))).)).))))))).))))))))..... (-28.68 = -29.44 + 0.76)

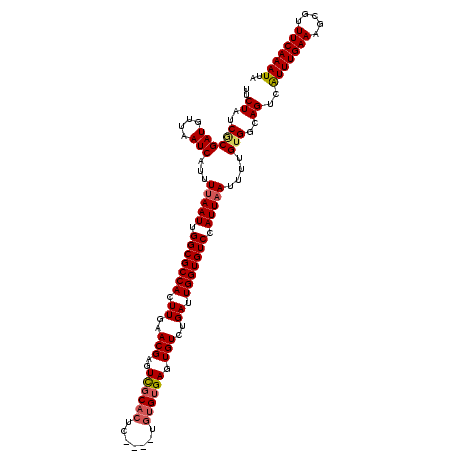
| Location | 14,403,562 – 14,403,678 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.02 |
| Mean single sequence MFE | -31.65 |
| Consensus MFE | -28.64 |
| Energy contribution | -29.08 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.57 |
| SVM RNA-class probability | 0.965002 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14403562 116 - 22407834 UUCUAUCGCGAUGUUAAUCAUUUUAAUUGGCGCCACUUGAACGAGUCGCACUC----UGUGUGAGUGUCUGAUUGGUGUCCAUUAAUUUUGUGGCAGUCAUUUGAAAGCGUUUCAAAUUA ......((((((....))).((((((.(((((((((..((((.((((((((((----.....))))))..)))).)).........))..))))).)))).))))))))).......... ( -32.40) >DroSec_CAF1 24666 116 - 1 UUCUAUCGCGAUGUUAAUCAUUUUAAUUGGCGCCACUUGAACGAGUCGCACUC----UGUGUGAGUGUCUGAUUGGUGUCCAUUAAUUUUGUGGCAGUCAUUUGAAAGCGUUUCAAAUUA ......((((((....))).((((((.(((((((((..((((.((((((((((----.....))))))..)))).)).........))..))))).)))).))))))))).......... ( -32.40) >DroSim_CAF1 23565 116 - 1 UUCUAUCGCGAUGUUAAUCAUUUUAAUUGGCGCCACUUGAACGAGUCGCACUC----UGUGUGAGUGUCUGAUUGGUGUCCAUUAAUUUUGUGGCAGUCAUUUGAAAGCGUUUCAAAUUA ......((((((....))).((((((.(((((((((..((((.((((((((((----.....))))))..)))).)).........))..))))).)))).))))))))).......... ( -32.40) >DroEre_CAF1 24767 116 - 1 UUCUAUCACGAUGUUAAUCAUUUUAAUUGGCGCCACUUGAACGAGUCGCACUC----UGUGUGAGUGUCUGAUUGGUGUCCAUUAAUUUUGUGGCAGUCAUUUGAAAGCGUUUCAAAUUA ..((..((((((....)))...(((((.(((((((.((..(((..((((((..----.)))))).)))..)).))))))).)))))....)))..))..(((((((.....))))))).. ( -30.70) >DroYak_CAF1 24744 116 - 1 UUCUAUCGCGAUGUUAAUCAUUUUAAUUGGCGCCACUUGAACGAGUCGCACUC----UGUGUGAGUGUCUGAUUGGUGUCCAUUAAUUUUGUGGCAGUCAUUUGAAAGCGUUUCAAAUUA ......((((((....))).((((((.(((((((((..((((.((((((((((----.....))))))..)))).)).........))..))))).)))).))))))))).......... ( -32.40) >DroAna_CAF1 38041 120 - 1 UUCUAUCGCGAUGUUAAUCAUUUUAAUUGGCGCCACUUGAACGGGUUUCUGUCGGUGUGUGCGAGUGUCUGAUUGGUGUCCAUUCAUUUUGGGGCAGUCGUUUGAAAGCGUUUCAAAUUA ......((((((....))).((((((...((((((((.((.(((....))))))))).))))((.(((((.(..((((......)))).).))))).))..))))))))).......... ( -29.60) >consensus UUCUAUCGCGAUGUUAAUCAUUUUAAUUGGCGCCACUUGAACGAGUCGCACUC____UGUGUGAGUGUCUGAUUGGUGUCCAUUAAUUUUGUGGCAGUCAUUUGAAAGCGUUUCAAAUUA ..((..((((((....)))...(((((.(((((((.((..(((..((((((.......)))))).)))..)).))))))).)))))....)))..))..(((((((.....))))))).. (-28.64 = -29.08 + 0.45)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:33:39 2006