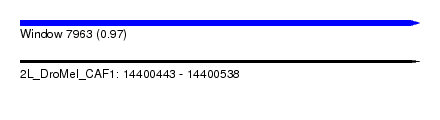
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,400,443 – 14,400,538 |
| Length | 95 |
| Max. P | 0.972993 |

| Location | 14,400,443 – 14,400,538 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.37 |
| Mean single sequence MFE | -19.36 |
| Consensus MFE | -17.57 |
| Energy contribution | -17.91 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.70 |
| SVM RNA-class probability | 0.972993 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
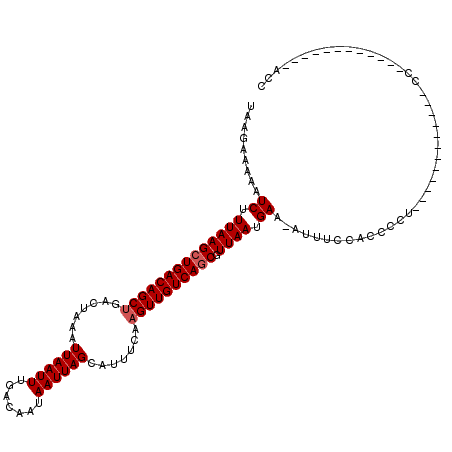
>2L_DroMel_CAF1 14400443 95 + 22407834 UAAGAAAAAUCUUUAAGCUGACAGCUGACUAAAUUAAUUUGACAAUAAUUAGCAUUUCAAGUUGUCAGCGUUAAUGAA-AUUUUCACCCCU------------CC------------ACC ...(((((.((.((((((((((((((.......((((((.......)))))).......)))))))))).)))).)).-.)))))......------------..------------... ( -21.84) >DroPse_CAF1 33163 84 + 1 UAUGAAAAAUCUUUAAGCUGACAGCUGACUAAAUUAAUUUGACAAUAAUUAGCAUUCCAAGUUGUCAACGUUAAUGAA-AUUUCU----------------------------------- ...((((..((.(((((.((((((((.......((((((.......)))))).......)))))))).).)))).)).-.)))).----------------------------------- ( -14.74) >DroSec_CAF1 21634 94 + 1 UAAGAAAAAUCUUUAAGCUGACAGCUGACUAAAUUAAUUUGACAAUAAUUAGCAUUUCAAGUUGUCAGCGUUAAUGA--AUUUCCACCCCU------------UC------------ACC ...((((..((.((((((((((((((.......((((((.......)))))).......)))))))))).)))).))--.)))).......------------..------------... ( -20.04) >DroEre_CAF1 21556 108 + 1 UAAGAAAAAUCUUUAAGCUGACAGCUGACUAAAUUAAUUUGACAAUAAUUAGCAUUUCAAGUUGUCAGCGUUAAUGAAAAUUUCCACACCUCCCGCCCUCCCUCC------------AAC ...((((..((.((((((((((((((.......((((((.......)))))).......)))))))))).)))).))...)))).....................------------... ( -20.24) >DroYak_CAF1 21537 97 + 1 UAAGAAAAAUCUUUAAGCUGACAGCUGACUAAAUUAAUUUGACAAUAAUUAGCAUUUCAAGUUGUCAGCGUUAAUGAA-AUUUCCACCCCUCCC----------C------------GCC ...((((..((.((((((((((((((.......((((((.......)))))).......)))))))))).)))).)).-.))))..........----------.------------... ( -21.14) >DroAna_CAF1 35863 111 + 1 UAAGAAGAAUCUUUAAGCUGACAGCCGACUAAAUUAAUUUGACAAUAAUUAGCAUUUCAAGUUGUCAGCGUUAAUGAA-AUC---GCCCCU-----CUACCCCCCAAGCAUCUCCUGCCC ..((.(((.((.(((((((((((((........((((((.......))))))........))))))))).)))).)).-...---((....-----...........)).))).)).... ( -18.15) >consensus UAAGAAAAAUCUUUAAGCUGACAGCUGACUAAAUUAAUUUGACAAUAAUUAGCAUUUCAAGUUGUCAGCGUUAAUGAA_AUUUCCACCCCU____________CC____________ACC .........((.((((((((((((((.......((((((.......)))))).......)))))))))).)))).))........................................... (-17.57 = -17.91 + 0.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:33:33 2006