| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,400,142 – 14,400,256 |
| Length | 114 |
| Max. P | 0.843262 |

| Location | 14,400,142 – 14,400,256 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 80.78 |
| Mean single sequence MFE | -16.23 |
| Consensus MFE | -11.15 |
| Energy contribution | -11.98 |
| Covariance contribution | 0.83 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.843262 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
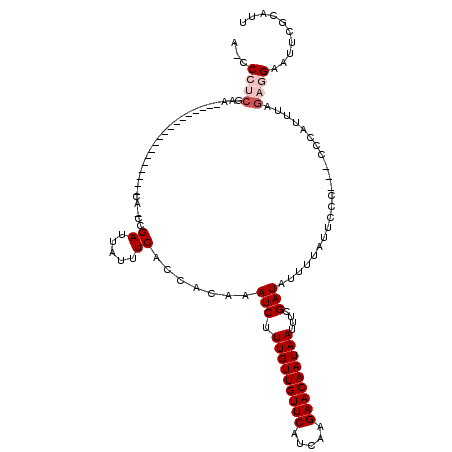
>2L_DroMel_CAF1 14400142 114 + 22407834 A-CCCUCGAACACACACACACUCGAAAACA-GCGAUUAUUUCACCACAAAUCUUUGUUGUUCAUCAAGAACAAUAAUUUCGAUAUUUUAUUCCC---CCCAUUUAGAGGAAUUCACAUA .-.(((((......)......((((((...-..((.....))...........(((((((((.....)))))))))))))))............---........)))).......... ( -17.30) >DroSec_CAF1 21351 96 + 1 A-CCCUCGAA------------------CA-GCGAUUAUUUCACCACAAAUCUUUGUUGUUCAUCAAGAACAAUAAUUUCGAUAUUUUAUUCCC---CCCAUUUAGAGGAAUUCGCAUU .-.(((((((------------------((-((((..((((......))))..))))))))).....(((.......)))..............---........)))).......... ( -17.60) >DroSim_CAF1 20232 96 + 1 A-CCCUCGAA------------------CA-GCGAUUAUUUCACCACAAAUCUUUGUUGUUCAUCAAGAACAAUAAUUUCGAUAUUUUAUUCCC---CCCAUUUAGAGGAAUUCGCAUU .-.(((((((------------------((-((((..((((......))))..))))))))).....(((.......)))..............---........)))).......... ( -17.60) >DroEre_CAF1 21260 110 + 1 GAACCUCGAACACACACG--------GCCA-CCGAUUAUUUCAGCACAAAUCUUUGUUGUUCAUCAAGAACAAUAAUUUCGAUAUUUUAUUCCCCCCCCCAUUUAGAGGAAUUCGCAUU (((((((.........((--------(...-)))...............(((.(((((((((.....)))))))))....)))......................))))..)))..... ( -16.80) >DroYak_CAF1 21253 92 + 1 A--C------------CG--------AGCG-CCGAUUAUUUCACCACAAAUCUUUGUUGUUCAUCAAGAACAAUAAUUUCGAUAUUUUAUUCC----CCCAUUUAGAGGAAUUCGCAUU .--.------------..--------.(((-..................(((.(((((((((.....)))))))))....))).....(((((----.(......).))))).)))... ( -14.80) >DroAna_CAF1 35580 96 + 1 A-UCCACA------------------GCCAUCCGAUUAUUUCACCACAAAUCUUUGUUGUUCGUCGAGAACAAUAAUUUUGAUACUUUAUUCC----CCCAUUUAGCAGAGCUCUCGUC .-.....(------------------(((....((.....))....((((...(((((((((.....))))))))).))))............----...........).)))...... ( -13.30) >consensus A_CCCUCGAA__________________CA_CCGAUUAUUUCACCACAAAUCUUUGUUGUUCAUCAAGAACAAUAAUUUCGAUAUUUUAUUCCC___CCCAUUUAGAGGAAUUCGCAUU ...((((..........................((.....)).......(((.(((((((((.....)))))))))....)))......................)))).......... (-11.15 = -11.98 + 0.83)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:33:32 2006