
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,393,644 – 14,393,781 |
| Length | 137 |
| Max. P | 0.961187 |

| Location | 14,393,644 – 14,393,748 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 88.22 |
| Mean single sequence MFE | -22.79 |
| Consensus MFE | -18.50 |
| Energy contribution | -18.34 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.52 |
| SVM RNA-class probability | 0.961187 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14393644 104 + 22407834 CUACAGUUA-UGUGCACAACUAAUGUUAAUAACAUUUAAUCAGCGCUCAUCAGGUUUUAUGGCCAUUUCCAAAUGAGCAAUUAAGCUAAUGAAUUUAUUAGCCAU ..(((....-)))((......(((((.....)))))......))((((((..((..............))..))))))......(((((((....)))))))... ( -18.54) >DroSec_CAF1 14652 104 + 1 CUACAGUUA-UGUGCACAACUAAUGUUAAUAACAUUUAAUCAGCGCUCAUCAGGUUUUAUGACCAUUUCCAAAUGAGCAAUUAGGCUAAUGAAAUUAUUAGCCAU ..(((....-)))((......(((((.....)))))......))((((((..((..............))..)))))).....((((((((....)))))))).. ( -22.74) >DroEre_CAF1 14764 105 + 1 CUGCGCUUAUUGUGUACAACUAAUGUUAAUAACAUUUAAUCAGCGCUCAUCAGGUUUUAUGGCCAUUUCCAAAUGAGCAAUUAGGCUAAUGAAAUUAUUAGCCAU ....(((((((((((......(((((.....)))))......))))......((((....)))).......))))))).....((((((((....)))))))).. ( -26.00) >DroYak_CAF1 14497 104 + 1 CUGCAGUUA-UGUGCACAGCUAAUGUUAAUAACAUUUAAUCAGCGCUCAUCAGGUUUUAUAGCCAUUUCCAAAUGAGCAAUUAGGCUAAUGAAAUUAUUAGCCAU .((((....-..))))..(((...(((((......))))).)))((((((..((..............))..)))))).....((((((((....)))))))).. ( -25.14) >DroAna_CAF1 29859 104 + 1 CUAUAAUCG-UAACCAGAACUCAUGCUAAUAGUAUUUAAUCAGCGUUCAUCGGAUUUUACGACCAUUUCUAGAUGAGCAAUUAGGCUAAUGAAAUUAUUAGCCGC .........-......((....((((.....))))....)).((((((((((((.............))).))))))).....((((((((....)))))))))) ( -21.52) >consensus CUACAGUUA_UGUGCACAACUAAUGUUAAUAACAUUUAAUCAGCGCUCAUCAGGUUUUAUGGCCAUUUCCAAAUGAGCAAUUAGGCUAAUGAAAUUAUUAGCCAU .............((......(((((.....)))))......))((((((..((..............))..)))))).....((((((((....)))))))).. (-18.50 = -18.34 + -0.16)

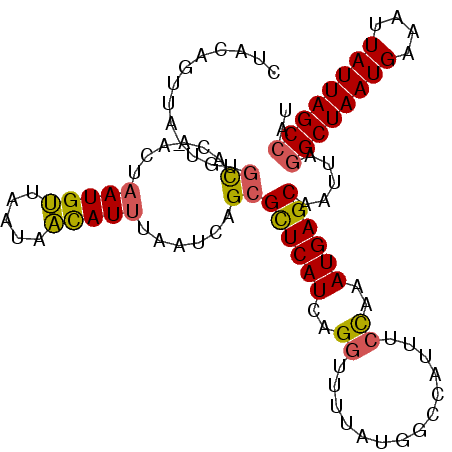
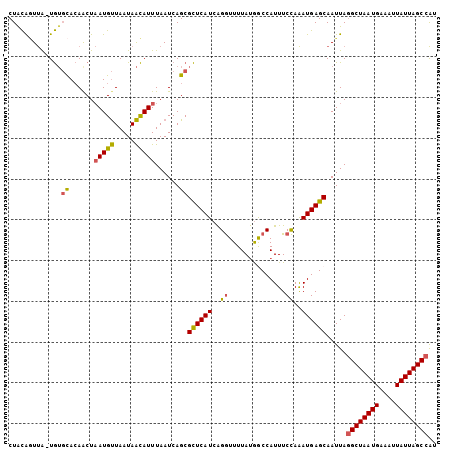
| Location | 14,393,644 – 14,393,748 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 88.22 |
| Mean single sequence MFE | -24.30 |
| Consensus MFE | -17.76 |
| Energy contribution | -17.72 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.733973 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14393644 104 - 22407834 AUGGCUAAUAAAUUCAUUAGCUUAAUUGCUCAUUUGGAAAUGGCCAUAAAACCUGAUGAGCGCUGAUUAAAUGUUAUUAACAUUAGUUGUGCACA-UAACUGUAG (((((((.....((((..(((......)))....))))..)))))))........(((.((((.((((((.(((.....))))))))))))).))-)........ ( -21.50) >DroSec_CAF1 14652 104 - 1 AUGGCUAAUAAUUUCAUUAGCCUAAUUGCUCAUUUGGAAAUGGUCAUAAAACCUGAUGAGCGCUGAUUAAAUGUUAUUAACAUUAGUUGUGCACA-UAACUGUAG ..(((((((......))))))).....(((((((.((..............)).)))))))(((((((((.(((.....)))))))))).))...-......... ( -23.94) >DroEre_CAF1 14764 105 - 1 AUGGCUAAUAAUUUCAUUAGCCUAAUUGCUCAUUUGGAAAUGGCCAUAAAACCUGAUGAGCGCUGAUUAAAUGUUAUUAACAUUAGUUGUACACAAUAAGCGCAG ..(((((((......))))))).....(((((((.((..............)).)))))))(((.....(((((.....))))).((((....)))).))).... ( -24.84) >DroYak_CAF1 14497 104 - 1 AUGGCUAAUAAUUUCAUUAGCCUAAUUGCUCAUUUGGAAAUGGCUAUAAAACCUGAUGAGCGCUGAUUAAAUGUUAUUAACAUUAGCUGUGCACA-UAACUGCAG ..(((((((......))))))).....(((((((.((..............)).)))))))(((((((((......)))..))))))..((((..-....)))). ( -25.34) >DroAna_CAF1 29859 104 - 1 GCGGCUAAUAAUUUCAUUAGCCUAAUUGCUCAUCUAGAAAUGGUCGUAAAAUCCGAUGAACGCUGAUUAAAUACUAUUAGCAUGAGUUCUGGUUA-CGAUUAUAG ..(((((((......))))))).................((((((((((...(((..((((((((((........))))))....))))))))))-))))))).. ( -25.90) >consensus AUGGCUAAUAAUUUCAUUAGCCUAAUUGCUCAUUUGGAAAUGGCCAUAAAACCUGAUGAGCGCUGAUUAAAUGUUAUUAACAUUAGUUGUGCACA_UAACUGUAG ..(((((((......))))))).....(((((((.((..............)).)))))))(((((((((......)))..)))))).................. (-17.76 = -17.72 + -0.04)

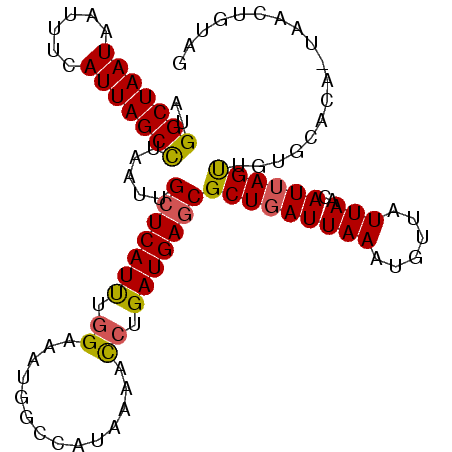
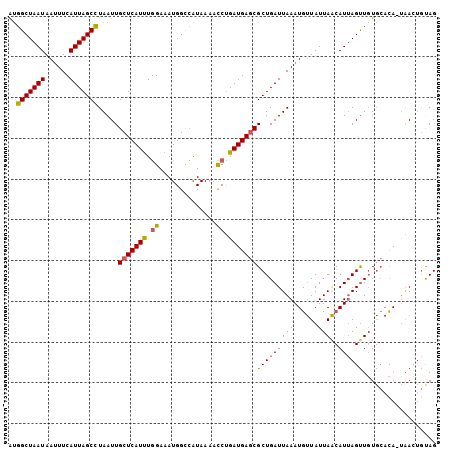
| Location | 14,393,682 – 14,393,781 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 88.38 |
| Mean single sequence MFE | -23.13 |
| Consensus MFE | -18.68 |
| Energy contribution | -18.76 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.906386 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14393682 99 + 22407834 UCAGCGCUCAUCAGGUUUUAUGGCCAUUUCCAAAUGAGCAAUUAAGCUAAUGAAUUUAUUAGCCAUUCGCGAUGUAUUGUGCAAUCAACAAAUGCCAAA ...((((.((((.((((....))))............(((((...(((((((....))))))).))).))))))....))))................. ( -20.40) >DroSec_CAF1 14690 99 + 1 UCAGCGCUCAUCAGGUUUUAUGACCAUUUCCAAAUGAGCAAUUAGGCUAAUGAAAUUAUUAGCCAUUCGCGAUGUAUUGUGCAAUAAACAAGUGCCAAA ...((((.((((.((((....))))............((.....((((((((....))))))))....))))))....))))................. ( -24.40) >DroEre_CAF1 14803 99 + 1 UCAGCGCUCAUCAGGUUUUAUGGCCAUUUCCAAAUGAGCAAUUAGGCUAAUGAAAUUAUUAGCCAUUCGUGAUAUAUUGUGCAAUAAACAAGUGCCAGA ...((((..((((.(.......((((((....)))).)).....((((((((....))))))))...).))))...((((.......)))))))).... ( -22.70) >DroYak_CAF1 14535 99 + 1 UCAGCGCUCAUCAGGUUUUAUAGCCAUUUCCAAAUGAGCAAUUAGGCUAAUGAAAUUAUUAGCCAUUCGUUAUAUAUUGUGCCAUAAAAAAAUGCCAAA ..(((((((((..((..............))..)))))).....((((((((....))))))))....))).....(((.((.((......))))))). ( -21.14) >DroAna_CAF1 29897 99 + 1 UCAGCGUUCAUCGGAUUUUACGACCAUUUCUAGAUGAGCAAUUAGGCUAAUGAAAUUAUUAGCCGCUUGUGAUAUACGGCGCAAUAAACAAGUGCUAUG ..(((((((((((((.............))).))))))).....((((((((....)))))))))))..........(((((.........)))))... ( -27.02) >consensus UCAGCGCUCAUCAGGUUUUAUGGCCAUUUCCAAAUGAGCAAUUAGGCUAAUGAAAUUAUUAGCCAUUCGUGAUAUAUUGUGCAAUAAACAAGUGCCAAA ...((((((((..((..............))..)))))).....((((((((....))))))))................))................. (-18.68 = -18.76 + 0.08)


| Location | 14,393,682 – 14,393,781 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 88.38 |
| Mean single sequence MFE | -22.31 |
| Consensus MFE | -20.08 |
| Energy contribution | -19.84 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.678194 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14393682 99 - 22407834 UUUGGCAUUUGUUGAUUGCACAAUACAUCGCGAAUGGCUAAUAAAUUCAUUAGCUUAAUUGCUCAUUUGGAAAUGGCCAUAAAACCUGAUGAGCGCUGA .(..((((((((.(((..........)))))))))(((((((......))))))).....(((((((.((..............)).)))))))))..) ( -22.14) >DroSec_CAF1 14690 99 - 1 UUUGGCACUUGUUUAUUGCACAAUACAUCGCGAAUGGCUAAUAAUUUCAUUAGCCUAAUUGCUCAUUUGGAAAUGGUCAUAAAACCUGAUGAGCGCUGA .(..((.........((((..........))))..(((((((......))))))).....(((((((.((..............)).)))))))))..) ( -24.04) >DroEre_CAF1 14803 99 - 1 UCUGGCACUUGUUUAUUGCACAAUAUAUCACGAAUGGCUAAUAAUUUCAUUAGCCUAAUUGCUCAUUUGGAAAUGGCCAUAAAACCUGAUGAGCGCUGA ...(((.............................(((((((......))))))).....(((((((.((..............)).)))))))))).. ( -22.04) >DroYak_CAF1 14535 99 - 1 UUUGGCAUUUUUUUAUGGCACAAUAUAUAACGAAUGGCUAAUAAUUUCAUUAGCCUAAUUGCUCAUUUGGAAAUGGCUAUAAAACCUGAUGAGCGCUGA .(..((..(((.(((((........))))).))).(((((((......))))))).....(((((((.((..............)).)))))))))..) ( -22.74) >DroAna_CAF1 29897 99 - 1 CAUAGCACUUGUUUAUUGCGCCGUAUAUCACAAGCGGCUAAUAAUUUCAUUAGCCUAAUUGCUCAUCUAGAAAUGGUCGUAAAAUCCGAUGAACGCUGA ..((((....((((((((.................(((((((......)))))))...((((....(((....)))..))))....)))))))))))). ( -20.60) >consensus UUUGGCACUUGUUUAUUGCACAAUAUAUCACGAAUGGCUAAUAAUUUCAUUAGCCUAAUUGCUCAUUUGGAAAUGGCCAUAAAACCUGAUGAGCGCUGA ..((((.............................(((((((......))))))).....(((((((.((..............)).))))))))))). (-20.08 = -19.84 + -0.24)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:33:26 2006