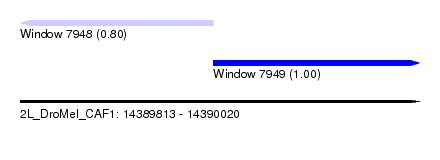
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,389,813 – 14,390,020 |
| Length | 207 |
| Max. P | 0.996017 |

| Location | 14,389,813 – 14,389,913 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 73.78 |
| Mean single sequence MFE | -42.32 |
| Consensus MFE | -15.36 |
| Energy contribution | -17.45 |
| Covariance contribution | 2.09 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.36 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.796111 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14389813 100 - 22407834 AUUAUACGACG--------AAGGAGCGG------GCCUUGGUGAA--GGGAACACCCUAUGACCUGGCCAGGCCAGGGCACGGUUCAGGCUCCGAC--UCUGGUCACCAUCUCUGUCC .......((((--------.(((....(------(((..(((.((--(((....)))).).))).)))).((((((((..(((........))).)--)))))))....))).)))). ( -39.20) >DroSec_CAF1 10869 100 - 1 AUUAUACGACG--------GAGGAGCGG------GCCUGGGAGAA--GGGAACACCCUAUGACCUGGCCAGGCCAGGCCACGGUUCAGGCUCCGAC--UCUGGUCACCAUCUCUGUCC .......((((--------(((((((((------(((((.....(--(((....))))..(((((((((......))))).))))))))).))).)--))(((...))).))))))). ( -46.40) >DroEre_CAF1 10727 101 - 1 AUUAUACGGCGC------AGGGGGGUGG------CGCUGCGAGAA--GGGAACACCCUUUGACCUGGCC-GGCCAGGCCAUGGUUCGGGCUCCGAU--UCUGGUCACCAUCUCUGUCC .......((.((------((((((((((------((((.((((((--(((....)))))).((((((((-.....))))).)))))))))..((..--..)))))))).))))))))) ( -42.80) >DroYak_CAF1 10663 102 - 1 AUUAUACGGCG--------GGGGAGUGG------GACUGGGAGAACUGGGUACACCCUCUGACCUGGCCAGGCCAGGCCAUGGUUCAGACUCCGAC--UCUGGUCACCAUCUCUGUCC .......((((--------(((..((((------.....((((....(((....)))((((((((((((......))))).)).)))))))))(((--....))).))))))))))). ( -43.60) >DroAna_CAF1 26113 112 - 1 AUUAUACGACUCGAGGCCAGAGGGACAGGACGAACACUGGGGGAG--CCGAACACACUCUGACCUGGCCUGGU----CUAUGGUUCAGACUCCGACUUUCUGGUCACCAUCUCUGGCC ..............(((((((((..(((........)))((((((--(.((((.((....((((......)))----)..)))))).).))))((((....)))).)).))))))))) ( -42.60) >DroPer_CAF1 16698 90 - 1 AUUAUAUGGCUC--------------CC-----ACACUG-UGGAG--GGGAGCAAACUCUGACCUGGCCAGG----GCUCUGGUUCAGGCUCUGGC--UACGGUCACCAUCUCUUUCC ........((((--------------((-----.(....-.)...--))))))......(((((((((((((----((.(((...)))))))))))--)).)))))............ ( -39.30) >consensus AUUAUACGACG________GAGGAGCGG______CACUGGGAGAA__GGGAACACCCUCUGACCUGGCCAGGCCAGGCCAUGGUUCAGGCUCCGAC__UCUGGUCACCAUCUCUGUCC .......(((...........(((((.....................(((....))).(((((((((((......))))).)).)))))))))(((......))).........))). (-15.36 = -17.45 + 2.09)

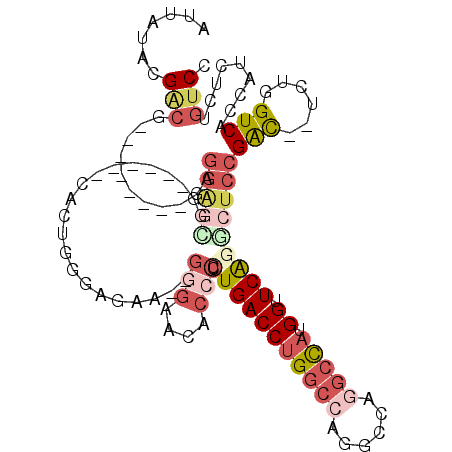
| Location | 14,389,913 – 14,390,020 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 69.81 |
| Mean single sequence MFE | -24.82 |
| Consensus MFE | -14.15 |
| Energy contribution | -14.65 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.57 |
| SVM decision value | 2.64 |
| SVM RNA-class probability | 0.996017 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14389913 107 + 22407834 ACAAUAAAUUGUCAGGUGGGGGGGCUAGCUGUCAAAAUGCAGGGGCAUAACUCUCUCUC------UUCUCCACCAACCCCCAACCUUGCCACCAAUCCCACCUUACAACCCCA- ........((((.((((((((((((..((.........)).((((..............------............))))......))).))...))))))).)))).....- ( -28.87) >DroPse_CAF1 14738 84 + 1 ACAAUAAAUUGUCAGGUAGGGCGGCUAGCUGUCAAAAUGCAAGGGCAUAACUCUCUUUC------UCCACCGCCCCCCC-CUGUCCU----------------------GCCU- .............(((((((((((...((.........))..((((.............------......))))....-)))))))----------------------))))- ( -23.91) >DroSec_CAF1 10969 104 + 1 ACAAUAAAUUGUCAGGUGGGGCGGCUAGCUGUCAAAAUGCAGGGGCAUAACUCUCUCUCUCGGCCUUCUCCACCAUCCC----------CACCAUACCCACCUUACCACCCAAC ..............(((((((.((....((((......))))((((................))))......))..)))----------))))..................... ( -22.79) >DroPer_CAF1 16788 85 + 1 ACAAUAAAUUGUCAGGUAGGGCGGCUAGCUGUCAAAAUGCAAGGGCAUAACUCUCUUUC------UCCGCCGCCCCCCCUCUGUCCU----------------------GCCU- .............(((((((((((...((.........))..((((.............------......)))).....)))))))----------------------))))- ( -23.71) >consensus ACAAUAAAUUGUCAGGUAGGGCGGCUAGCUGUCAAAAUGCAAGGGCAUAACUCUCUCUC______UCCUCCACCACCCC_CUGUCCU______________________CCCA_ ..............((((((((((....)))))...((((....)))).....................)))))........................................ (-14.15 = -14.65 + 0.50)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:33:20 2006