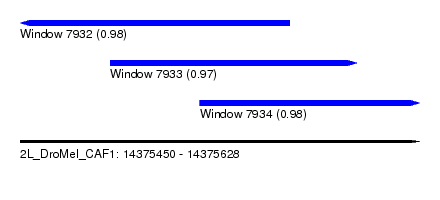
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,375,450 – 14,375,628 |
| Length | 178 |
| Max. P | 0.983415 |

| Location | 14,375,450 – 14,375,570 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.81 |
| Mean single sequence MFE | -24.09 |
| Consensus MFE | -21.38 |
| Energy contribution | -21.62 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.94 |
| SVM RNA-class probability | 0.983415 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14375450 120 - 22407834 UAUAUGUCAAACUGGUGAAUAAAUCAAUUUGACAGCUAAACGCUGCUGUUGAAAGACAGAGAGGCCAAUAGAAAGACUGUCAGAAAAAAGCGGGUAAAAAGAGAGUUGUAUAAAUCAAAG ....(((((((.((((......)))).))))))).(((...(((.(((((....)))))...)))...)))...((((.((...................)).))))............. ( -24.91) >DroSec_CAF1 23624 119 - 1 UAUAUGUCAAACUGGUGAAUAAAUCAAUUUGACAGCUAAACGCUGCUGUUGAAAGACAGAGAGGCCAAUAGAAAGACAGCCAGGA-AAUGCGAGUAAGAAGAGAGUUGUAUAAAUCAAAG ....(((((((.((((......)))).))))))).(((...(((.(((((....)))))...)))...)))..............-.((((((.(........).))))))......... ( -24.90) >DroSim_CAF1 25726 119 - 1 UAUAUGUCAAACUGGUGAAUAAAUCAAUUUGACAGCUAAACGCUGCUGUUGAAAGACAGAGAGGCCAAUAGAAAGACAGCCAGGA-AAUGCGAGUAAGAAGAGAGUUGUAUAAAUCAAAG ....(((((((.((((......)))).))))))).(((...(((.(((((....)))))...)))...)))..............-.((((((.(........).))))))......... ( -24.90) >DroEre_CAF1 24364 119 - 1 UAUAUGUCAAACUGGCGAAUAAAUCAAUUUGACAGCUAAACGCUGCUGUUGAAAGACAGAGAGGCAAAUAGAAAGACAGUCAGGA-AAUGCGGGUAAGAAAAGAGUUGUAUAAAUCAAAU ...........(((((.......((.(((((.((((.....))))(((((....))))).....))))).))......)))))..-.((((((.(........).))))))......... ( -23.02) >DroYak_CAF1 25210 119 - 1 UAUAUGUCAAACUGGCGAAUAAAUCAAUUUGACAGCUGAACGCUGCUGUUGAAAGACAGAGAGCCCAAUAGAAAGACAGUCAGGA-AAUGCGGGUAAGAAGAGAGUUGUAUAAGUCAAAG (((.((((.....)))).)))......((((((..((((..(((.(((((....)))))..)))......(.....)..))))..-.((((((.(........).))))))..)))))). ( -22.70) >consensus UAUAUGUCAAACUGGUGAAUAAAUCAAUUUGACAGCUAAACGCUGCUGUUGAAAGACAGAGAGGCCAAUAGAAAGACAGUCAGGA_AAUGCGGGUAAGAAGAGAGUUGUAUAAAUCAAAG ....(((((((.((((......)))).))))))).(((...(((.(((((....)))))...)))...)))...........((...((((((.(........).))))))...)).... (-21.38 = -21.62 + 0.24)

| Location | 14,375,490 – 14,375,600 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 96.73 |
| Mean single sequence MFE | -26.68 |
| Consensus MFE | -23.78 |
| Energy contribution | -23.82 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.970612 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14375490 110 + 22407834 ACAGUCUUUCUAUUGGCCUCUCUGUCUUUCAACAGCAGCGUUUAGCUGUCAAAUUGAUUUAUUCACCAGUUUGACAUAUAUUAAUGGUCGUUGGUGUAAAAUAAACGCGC ...(((........)))....((((......))))..(((((((..((((((((((..........))))))))))(((((((((....)))))))))...))))))).. ( -27.10) >DroSec_CAF1 23663 110 + 1 GCUGUCUUUCUAUUGGCCUCUCUGUCUUUCAACAGCAGCGUUUAGCUGUCAAAUUGAUUUAUUCACCAGUUUGACAUAUAUUAAUGGUCGUUGGUGUAAAAUAAAUGCGC (((((.........(((......))).....))))).(((((((..((((((((((..........))))))))))(((((((((....)))))))))...))))))).. ( -28.24) >DroSim_CAF1 25765 110 + 1 GCUGUCUUUCUAUUGGCCUCUCUGUCUUUCAACAGCAGCGUUUAGCUGUCAAAUUGAUUUAUUCACCAGUUUGACAUAUAUUAAUGGUCGUUGGUGUAAAAUAAAUGCGC (((((.........(((......))).....))))).(((((((..((((((((((..........))))))))))(((((((((....)))))))))...))))))).. ( -28.24) >DroEre_CAF1 24403 110 + 1 ACUGUCUUUCUAUUUGCCUCUCUGUCUUUCAACAGCAGCGUUUAGCUGUCAAAUUGAUUUAUUCGCCAGUUUGACAUAUAUUAAUGGCCGUUGGUGUAAAAUAAAUGCGC .....................((((......))))..(((((((..((((((((((..........))))))))))(((((((((....)))))))))...))))))).. ( -23.90) >DroYak_CAF1 25249 110 + 1 ACUGUCUUUCUAUUGGGCUCUCUGUCUUUCAACAGCAGCGUUCAGCUGUCAAAUUGAUUUAUUCGCCAGUUUGACAUAUAUUAAUGGUCGUUGGUGUAAAAUAAAUGCGC .............(((((.((((((......)))).)).)))))((((((((((((..........))))))))))(((((((((....)))))))))........)).. ( -25.90) >consensus ACUGUCUUUCUAUUGGCCUCUCUGUCUUUCAACAGCAGCGUUUAGCUGUCAAAUUGAUUUAUUCACCAGUUUGACAUAUAUUAAUGGUCGUUGGUGUAAAAUAAAUGCGC .....................((((......))))..(((((((..((((((((((..........))))))))))(((((((((....)))))))))...))))))).. (-23.78 = -23.82 + 0.04)

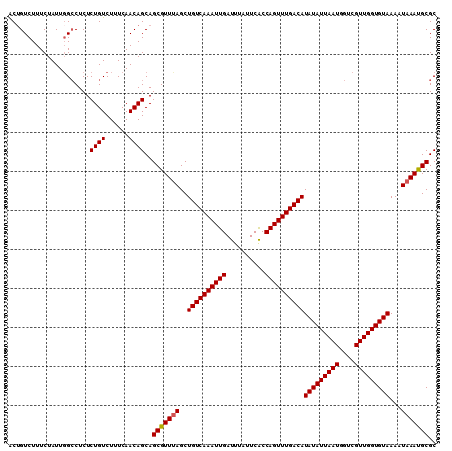
| Location | 14,375,530 – 14,375,628 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 97.55 |
| Mean single sequence MFE | -20.38 |
| Consensus MFE | -18.34 |
| Energy contribution | -19.14 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.975836 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14375530 98 + 22407834 UUUAGCUGUCAAAUUGAUUUAUUCACCAGUUUGACAUAUAUUAAUGGUCGUUGGUGUAAAAUAAACGCGCAUUAAAUUAUAAAUAUGUGAAGCUUAUU ...(((((((((((((..........)))))))))(((((((.(((((...((((((.......)))).))....))))).)))))))..)))).... ( -21.10) >DroSec_CAF1 23703 98 + 1 UUUAGCUGUCAAAUUGAUUUAUUCACCAGUUUGACAUAUAUUAAUGGUCGUUGGUGUAAAAUAAAUGCGCAUUAAAUUAUAAAUAUGUGAAGCUUAUU ...(((((((((((((..........)))))))))(((((((.(((((.....(((((.......))))).....))))).)))))))..)))).... ( -21.90) >DroSim_CAF1 25805 98 + 1 UUUAGCUGUCAAAUUGAUUUAUUCACCAGUUUGACAUAUAUUAAUGGUCGUUGGUGUAAAAUAAAUGCGCAUUAAAUUAUAAAUAUGUGAAGCUUAUU ...(((((((((((((..........)))))))))(((((((.(((((.....(((((.......))))).....))))).)))))))..)))).... ( -21.90) >DroEre_CAF1 24443 98 + 1 UUUAGCUGUCAAAUUGAUUUAUUCGCCAGUUUGACAUAUAUUAAUGGCCGUUGGUGUAAAAUAAAUGCGCAUUAAAUUAUAAAUAUGUGAAUCUUAUU ......((((((((((..........))))))))))(((((((((....))))))))).(((((...(((((............)))))....))))) ( -17.90) >DroYak_CAF1 25289 98 + 1 UUCAGCUGUCAAAUUGAUUUAUUCGCCAGUUUGACAUAUAUUAAUGGUCGUUGGUGUAAAAUAAAUGCGCAUUAAAUUAUAAAUAUGUGAAUCUUAUU ((((..((((((((((..........))))))))))((((((.(((((.....(((((.......))))).....))))).))))))))))....... ( -19.10) >consensus UUUAGCUGUCAAAUUGAUUUAUUCACCAGUUUGACAUAUAUUAAUGGUCGUUGGUGUAAAAUAAAUGCGCAUUAAAUUAUAAAUAUGUGAAGCUUAUU ...(((((((((((((..........)))))))))(((((((.(((((.....(((((.......))))).....))))).)))))))..)))).... (-18.34 = -19.14 + 0.80)

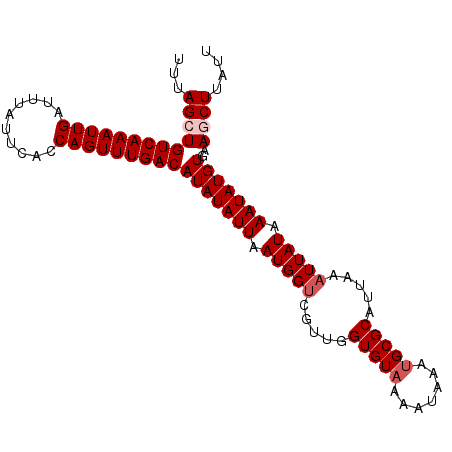
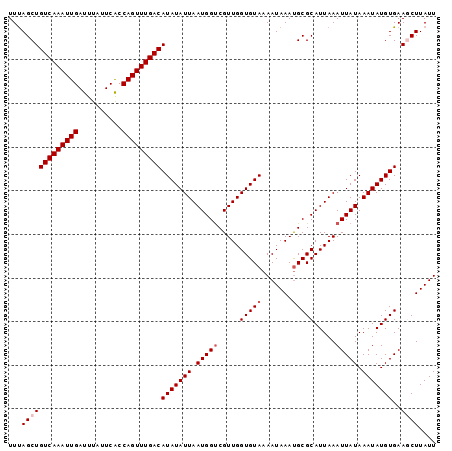
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:33:05 2006