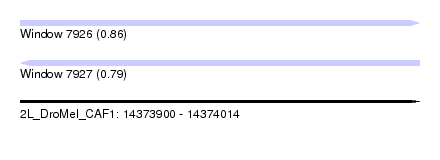
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,373,900 – 14,374,014 |
| Length | 114 |
| Max. P | 0.857847 |

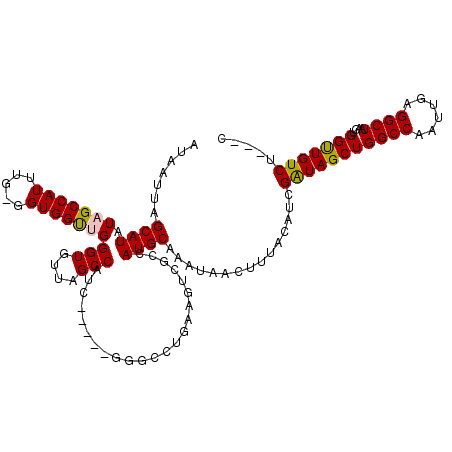
| Location | 14,373,900 – 14,374,014 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.30 |
| Mean single sequence MFE | -29.14 |
| Consensus MFE | -17.86 |
| Energy contribution | -19.36 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.59 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.857847 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14373900 114 + 22407834 GAGCAGACAACCACUUAGCCUCAAUUGGCCAGCUAUCGAUGUAAAGUUAUUUGCAUGGCGACUUCAAUCCC-----GAUGGCUAACACCCAACCACC-CAAAUGGCUAUAUGCUAAUUAU .((((.....(((....(((......))).(((((((.(((((((....)))))))((.((......))))-----)))))))..............-....))).....))))...... ( -27.70) >DroSec_CAF1 22024 109 + 1 -----GACAACCACUUAGCCUCAAUUGGCCAGCUAUCGAUGUAAAGUUAUUUGCAUGGCGACUUCAGGCCC-----GAUGGCUAUCACCCAACCACC-CAAAUGGCUAUAUGCUAAUUAU -----............(((......))).(((((((((((((((....)))))))(((........))))-----)))))))..............-....((((.....))))..... ( -27.70) >DroSim_CAF1 24125 110 + 1 -----GACAACCACUUAGCCUCAAUUGGCCAGCUAUCGAUGUAAAGUUAUUUGCAUGGCGACUUCAGGCCC-----GAUGGCUAUCACCCAACCACCCCAAAUGGCUAUAUGCUAAUUAU -----............(((......))).(((((((((((((((....)))))))(((........))))-----)))))))...................((((.....))))..... ( -27.70) >DroEre_CAF1 22801 97 + 1 GUAUAGACAGCCACUUAGCCCCAAUUGGCCAGCUAUCGAUGUAAAGUUAUUUGCAUU----------------------GGCUAACACCCCACCACC-CAAAUGGCUAUAUGCUAAUUAU (((((...(((((....(((......))).(((...(((((((((....))))))))----------------------))))..............-....))))).)))))....... ( -23.30) >DroYak_CAF1 23582 119 + 1 GAAUAGACAGCCACUUAGCCUCAAUUGGCCAGCUACCGAUGUAAAGUUAUUUGCAUUGCGAUUUCAAGCUGGCUAAGAUGGCUAACACCCAGCCACC-CAAAUGGCAAUAUGCUAAUUAU ........(((...((((((....((((((((((..(((((((((....))))))))).(....).))))))))))...))))))......((((..-....)))).....)))...... ( -39.30) >consensus G___AGACAACCACUUAGCCUCAAUUGGCCAGCUAUCGAUGUAAAGUUAUUUGCAUGGCGACUUCAAGCCC_____GAUGGCUAACACCCAACCACC_CAAAUGGCUAUAUGCUAAUUAU .................(((......))).(((((((.(((((((....)))))))(((........)))......)))))))...................((((.....))))..... (-17.86 = -19.36 + 1.50)

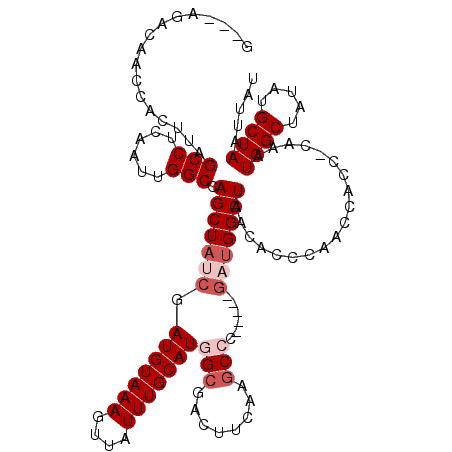
| Location | 14,373,900 – 14,374,014 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.30 |
| Mean single sequence MFE | -34.47 |
| Consensus MFE | -24.50 |
| Energy contribution | -24.34 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.787752 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14373900 114 - 22407834 AUAAUUAGCAUAUAGCCAUUUG-GGUGGUUGGGUGUUAGCCAUC-----GGGAUUGAAGUCGCCAUGCAAAUAACUUUACAUCGAUAGCUGGCCAAUUGAGGCUAAGUGGUUGUCUGCUC ......((((.(((((((((((-(.(.(((((...(((((.(((-----((...((((((.............))))))..))))).))))))))))...).)))))))))))).)))). ( -36.22) >DroSec_CAF1 22024 109 - 1 AUAAUUAGCAUAUAGCCAUUUG-GGUGGUUGGGUGAUAGCCAUC-----GGGCCUGAAGUCGCCAUGCAAAUAACUUUACAUCGAUAGCUGGCCAAUUGAGGCUAAGUGGUUGUC----- .......((...(((((((...-.))))))).))(((((((((.-----.(((((.((...((((.((...................))))))...)).)))))..)))))))))----- ( -36.51) >DroSim_CAF1 24125 110 - 1 AUAAUUAGCAUAUAGCCAUUUGGGGUGGUUGGGUGAUAGCCAUC-----GGGCCUGAAGUCGCCAUGCAAAUAACUUUACAUCGAUAGCUGGCCAAUUGAGGCUAAGUGGUUGUC----- .......((...(((((((.....))))))).))(((((((((.-----.(((((.((...((((.((...................))))))...)).)))))..)))))))))----- ( -36.51) >DroEre_CAF1 22801 97 - 1 AUAAUUAGCAUAUAGCCAUUUG-GGUGGUGGGGUGUUAGCC----------------------AAUGCAAAUAACUUUACAUCGAUAGCUGGCCAAUUGGGGCUAAGUGGCUGUCUAUAC .....((((((...(((((...-.)))))...))))))((.----------------------...))...............(((((((((((......))))....)))))))..... ( -26.10) >DroYak_CAF1 23582 119 - 1 AUAAUUAGCAUAUUGCCAUUUG-GGUGGCUGGGUGUUAGCCAUCUUAGCCAGCUUGAAAUCGCAAUGCAAAUAACUUUACAUCGGUAGCUGGCCAAUUGAGGCUAAGUGGCUGUCUAUUC .....((((((...(((((...-.)))))...))))))((((.(((((((.(((((......))).))...............(((.....)))......)))))))))))......... ( -37.00) >consensus AUAAUUAGCAUAUAGCCAUUUG_GGUGGUUGGGUGUUAGCCAUC_____GGGCCUGAAGUCGCCAUGCAAAUAACUUUACAUCGAUAGCUGGCCAAUUGAGGCUAAGUGGUUGUCU___C .......((((.(((((((.....)))))))(((....))).......................))))...............(((((((((((......))))....)))))))..... (-24.50 = -24.34 + -0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:32:58 2006