
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,369,688 – 14,370,075 |
| Length | 387 |
| Max. P | 0.999940 |

| Location | 14,369,688 – 14,369,808 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.28 |
| Mean single sequence MFE | -37.53 |
| Consensus MFE | -32.10 |
| Energy contribution | -33.74 |
| Covariance contribution | 1.64 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.45 |
| Structure conservation index | 0.86 |
| SVM decision value | 4.09 |
| SVM RNA-class probability | 0.999791 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14369688 120 + 22407834 AACACUCGAGAGGGGCUAACACAAAUUUAACACUGGCCGCCGCCAAUGUAGCCAUCAUGCGGCAUGCAAUAUGCUCGACUAACGAGCAAAAACAACAGCAACGGCGGAGUGAAGUGAAGC ..(((((....(.(((((...............))))).)((((..(((((((.......))).))))...((((((.....))))))..............)))))))))......... ( -36.96) >DroSec_CAF1 17738 120 + 1 AACACUCGAGAGCGGCUAACACAAAUUUAACACUGGCCGCCGCCAAUGUAGCCAUCAUGCGGCAUGCAAUAUGCUCGACUAACGAGCAAAAACAACAGCAACGGCGGAGUGAAGUGAAGC ..(((((....(((((((...............)))))))((((..(((((((.......))).))))...((((((.....))))))..............)))))))))......... ( -41.06) >DroSim_CAF1 19839 120 + 1 AACACUCGAGAGCGGCUAACACAAAUUUAACACUGGCCGCCGCCAAUGUAGCCAUCAUGCGGCAUGCAAUAUGCUCGACUAACGAGCAAAAACAACAGCAACGGCGGAGUGAAGUGAAGC ..(((((....(((((((...............)))))))((((..(((((((.......))).))))...((((((.....))))))..............)))))))))......... ( -41.06) >DroEre_CAF1 18830 106 + 1 AACACUCGAGAGCGGCUAACACAAAUUUAACACUGGGUGCCGCCAAUGUAGCCAUCAUGUGGCAUGCAAUAUGCUCGACUAACGAGCAAAAACAACAGCAACAGC------A-------- .........(.(((((...((............))...))))))..(((((((((...))))).))))...((((((.....)))))).........((....))------.-------- ( -27.50) >DroYak_CAF1 19256 112 + 1 AACACUCGAGAGCGGCUAACACAAAUUUAACACUGGCCGCCGCCAAUGUAGCCAUCAUGCGGCAUGCAAUAUGCUCGACUAACGAGCAAAAACAACAGCAACGGCGGAGUGA-------- ..(((((....(((((((...............)))))))((((..(((((((.......))).))))...((((((.....))))))..............))))))))).-------- ( -41.06) >consensus AACACUCGAGAGCGGCUAACACAAAUUUAACACUGGCCGCCGCCAAUGUAGCCAUCAUGCGGCAUGCAAUAUGCUCGACUAACGAGCAAAAACAACAGCAACGGCGGAGUGAAGUGAAGC ..(((((....(((((((...............)))))))((((..(((((((.......))).))))...((((((.....))))))..............)))))))))......... (-32.10 = -33.74 + 1.64)


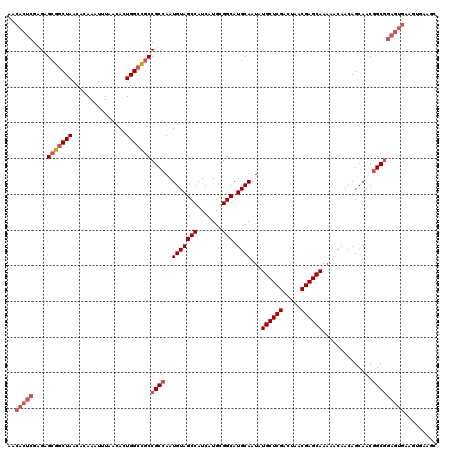
| Location | 14,369,688 – 14,369,808 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.28 |
| Mean single sequence MFE | -38.00 |
| Consensus MFE | -33.82 |
| Energy contribution | -35.10 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.90 |
| SVM RNA-class probability | 0.981938 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14369688 120 - 22407834 GCUUCACUUCACUCCGCCGUUGCUGUUGUUUUUGCUCGUUAGUCGAGCAUAUUGCAUGCCGCAUGAUGGCUACAUUGGCGGCGGCCAGUGUUAAAUUUGUGUUAGCCCCUCUCGAGUGUU .........(((((.((((((((.(((((...((((((.....))))))....))).)).)))..))))).((((((((....))))))))......................))))).. ( -38.70) >DroSec_CAF1 17738 120 - 1 GCUUCACUUCACUCCGCCGUUGCUGUUGUUUUUGCUCGUUAGUCGAGCAUAUUGCAUGCCGCAUGAUGGCUACAUUGGCGGCGGCCAGUGUUAAAUUUGUGUUAGCCGCUCUCGAGUGUU .........(((((((((((.(((((.((...((((((.....)))))).)).))).)).))..((((....))))))))(((((.((..(.......)..)).)))))....))))).. ( -40.70) >DroSim_CAF1 19839 120 - 1 GCUUCACUUCACUCCGCCGUUGCUGUUGUUUUUGCUCGUUAGUCGAGCAUAUUGCAUGCCGCAUGAUGGCUACAUUGGCGGCGGCCAGUGUUAAAUUUGUGUUAGCCGCUCUCGAGUGUU .........(((((((((((.(((((.((...((((((.....)))))).)).))).)).))..((((....))))))))(((((.((..(.......)..)).)))))....))))).. ( -40.70) >DroEre_CAF1 18830 106 - 1 --------U------GCUGUUGCUGUUGUUUUUGCUCGUUAGUCGAGCAUAUUGCAUGCCACAUGAUGGCUACAUUGGCGGCACCCAGUGUUAAAUUUGUGUUAGCCGCUCUCGAGUGUU --------.------((((.(((((((((...((((((.....))))))....))..((((.....))))......)))))))..))))(((((.......)))))((((....)))).. ( -29.20) >DroYak_CAF1 19256 112 - 1 --------UCACUCCGCCGUUGCUGUUGUUUUUGCUCGUUAGUCGAGCAUAUUGCAUGCCGCAUGAUGGCUACAUUGGCGGCGGCCAGUGUUAAAUUUGUGUUAGCCGCUCUCGAGUGUU --------.(((((((((((.(((((.((...((((((.....)))))).)).))).)).))..((((....))))))))(((((.((..(.......)..)).)))))....))))).. ( -40.70) >consensus GCUUCACUUCACUCCGCCGUUGCUGUUGUUUUUGCUCGUUAGUCGAGCAUAUUGCAUGCCGCAUGAUGGCUACAUUGGCGGCGGCCAGUGUUAAAUUUGUGUUAGCCGCUCUCGAGUGUU .........(((((.((((((((.(((((...((((((.....))))))....))).)).)))..)))))......((((((((((((........))).))).))))))...))))).. (-33.82 = -35.10 + 1.28)

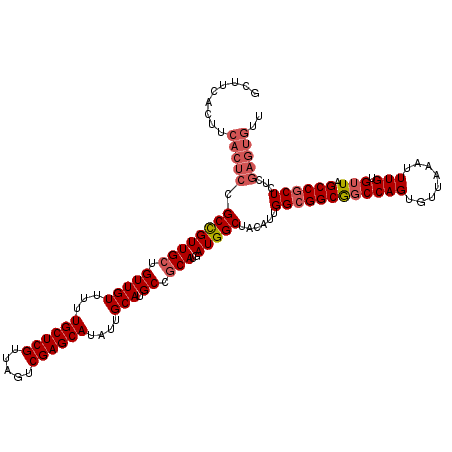
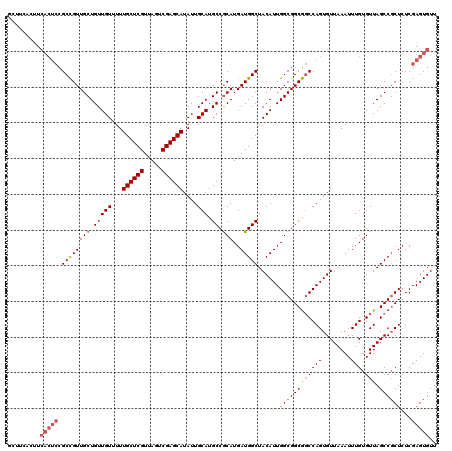
| Location | 14,369,728 – 14,369,848 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.08 |
| Mean single sequence MFE | -34.11 |
| Consensus MFE | -28.51 |
| Energy contribution | -28.55 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.84 |
| SVM decision value | 2.21 |
| SVM RNA-class probability | 0.990321 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14369728 120 + 22407834 CGCCAAUGUAGCCAUCAUGCGGCAUGCAAUAUGCUCGACUAACGAGCAAAAACAACAGCAACGGCGGAGUGAAGUGAAGCCGGAGCUGGAGUUGGAGUAACUGCAACAUGGAAACUACCA .......((((...(((((..((((((....((((((.....))))))..(((..((((..((((.............))))..))))..)))...)))..)))..)))))...)))).. ( -38.52) >DroSec_CAF1 17778 120 + 1 CGCCAAUGUAGCCAUCAUGCGGCAUGCAAUAUGCUCGACUAACGAGCAAAAACAACAGCAACGGCGGAGUGAAGUGAAGCCGGAGUUGGAGUUGGAGUAACUGCAACAUGGAAACUACCA .......((((...(((((..((((((....((((((.....))))))..(((..((((..((((.............))))..))))..)))...)))..)))..)))))...)))).. ( -36.12) >DroSim_CAF1 19879 120 + 1 CGCCAAUGUAGCCAUCAUGCGGCAUGCAAUAUGCUCGACUAACGAGCAAAAACAACAGCAACGGCGGAGUGAAGUGAAGCCGGAGUUGGAGUUGGAGUAACUGCAACAUGGAAACUACCA .......((((...(((((..((((((....((((((.....))))))..(((..((((..((((.............))))..))))..)))...)))..)))..)))))...)))).. ( -36.12) >DroEre_CAF1 18870 103 + 1 CGCCAAUGUAGCCAUCAUGUGGCAUGCAAUAUGCUCGACUAACGAGCAAAAACAACAGCAACAGC------A-----------AGUCGGAGUUGGAGUAACUGCAACAUGGAAACUACCA ..(((.(((.(((((...))))).((((...((((((.....)))))).........((..((((------.-----------.......))))..))...))))))))))......... ( -27.00) >DroYak_CAF1 19296 109 + 1 CGCCAAUGUAGCCAUCAUGCGGCAUGCAAUAUGCUCGACUAACGAGCAAAAACAACAGCAACGGCGGAGUGA-----------AGUUGGAGUUGGAGUAACUGCAACAUGGAAACUACCA .(((..(((((((.......))).))))...((((((.....))))))..............)))((.....-----------.((((.(((((...))))).))))..(....)..)). ( -32.80) >consensus CGCCAAUGUAGCCAUCAUGCGGCAUGCAAUAUGCUCGACUAACGAGCAAAAACAACAGCAACGGCGGAGUGAAGUGAAGCCGGAGUUGGAGUUGGAGUAACUGCAACAUGGAAACUACCA ..(((.(((.(((.......))).((((...((((((.....))))))...((..((((..((((...................))))..))))..))...))))))))))......... (-28.51 = -28.55 + 0.04)

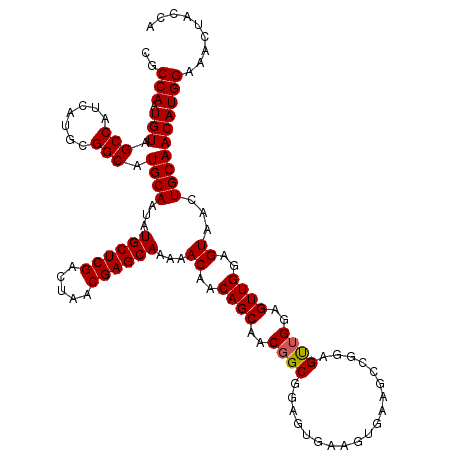
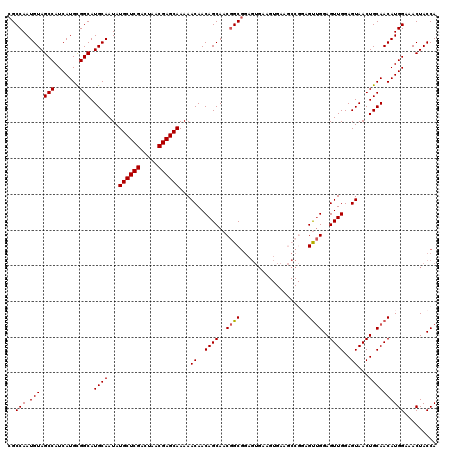
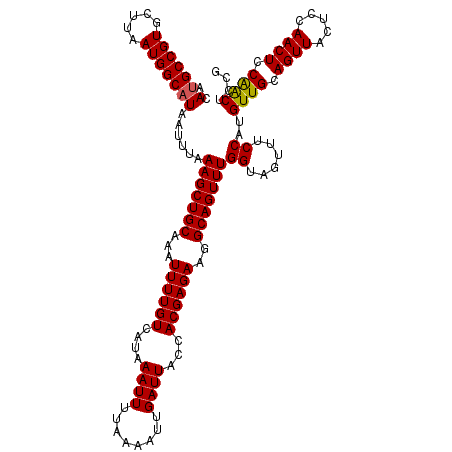
| Location | 14,369,728 – 14,369,848 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.08 |
| Mean single sequence MFE | -35.16 |
| Consensus MFE | -31.63 |
| Energy contribution | -31.15 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.90 |
| SVM decision value | 2.73 |
| SVM RNA-class probability | 0.996697 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14369728 120 - 22407834 UGGUAGUUUCCAUGUUGCAGUUACUCCAACUCCAGCUCCGGCUUCACUUCACUCCGCCGUUGCUGUUGUUUUUGCUCGUUAGUCGAGCAUAUUGCAUGCCGCAUGAUGGCUACAUUGGCG ..((((((..(((((.(((((......(((..((((..((((.............))))..))))..)))..((((((.....))))))....)).))).)))))..))))))....... ( -40.22) >DroSec_CAF1 17778 120 - 1 UGGUAGUUUCCAUGUUGCAGUUACUCCAACUCCAACUCCGGCUUCACUUCACUCCGCCGUUGCUGUUGUUUUUGCUCGUUAGUCGAGCAUAUUGCAUGCCGCAUGAUGGCUACAUUGGCG ..((((((..(((((.(((((.....((((..((((..(((............)))..))))..))))....((((((.....))))))....)).))).)))))..))))))....... ( -35.40) >DroSim_CAF1 19879 120 - 1 UGGUAGUUUCCAUGUUGCAGUUACUCCAACUCCAACUCCGGCUUCACUUCACUCCGCCGUUGCUGUUGUUUUUGCUCGUUAGUCGAGCAUAUUGCAUGCCGCAUGAUGGCUACAUUGGCG ..((((((..(((((.(((((.....((((..((((..(((............)))..))))..))))....((((((.....))))))....)).))).)))))..))))))....... ( -35.40) >DroEre_CAF1 18870 103 - 1 UGGUAGUUUCCAUGUUGCAGUUACUCCAACUCCGACU-----------U------GCUGUUGCUGUUGUUUUUGCUCGUUAGUCGAGCAUAUUGCAUGCCACAUGAUGGCUACAUUGGCG ..((((((..(((((.(((((.....((((..((((.-----------.------...))))..))))....((((((.....))))))....)).))).)))))..))))))....... ( -32.90) >DroYak_CAF1 19296 109 - 1 UGGUAGUUUCCAUGUUGCAGUUACUCCAACUCCAACU-----------UCACUCCGCCGUUGCUGUUGUUUUUGCUCGUUAGUCGAGCAUAUUGCAUGCCGCAUGAUGGCUACAUUGGCG ..((((((..(((((.(((((.....((((..((((.-----------..........))))..))))....((((((.....))))))....)).))).)))))..))))))....... ( -31.90) >consensus UGGUAGUUUCCAUGUUGCAGUUACUCCAACUCCAACUCCGGCUUCACUUCACUCCGCCGUUGCUGUUGUUUUUGCUCGUUAGUCGAGCAUAUUGCAUGCCGCAUGAUGGCUACAUUGGCG ..((((((..(((((.(((((.....((((..((((......................))))..))))....((((((.....))))))....)).))).)))))..))))))....... (-31.63 = -31.15 + -0.48)

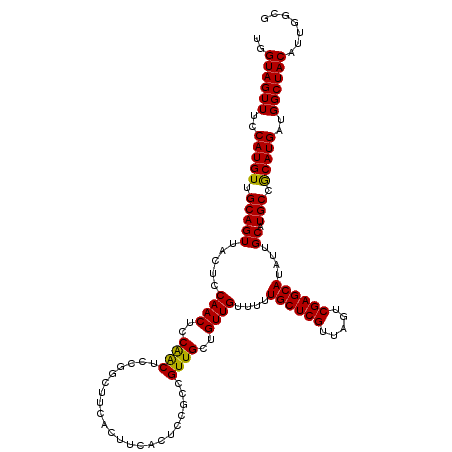
| Location | 14,369,808 – 14,369,928 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.16 |
| Mean single sequence MFE | -26.14 |
| Consensus MFE | -26.12 |
| Energy contribution | -26.00 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.21 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.820706 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14369808 120 - 22407834 ACAUGCCGUGCUUAAUGGCAUAAUUUAAAGCUGCAAAUUUUGUCAUAAAUUUUAAAAUUGAUUACCACGAGAAGGCAGUUUGGUAGUUUCCAUGUUGCAGUUACUCCAACUCCAGCUCCG ..(((((((.....)))))))......(((((((...((((((....((((........))))...))))))..)))))))((......))..((((.((((.....)))).)))).... ( -26.70) >DroSec_CAF1 17858 120 - 1 ACAUGCCGUGCUUAAUGGCAUAAUUUAAAGCUGCAAAUUUUGUCAUAAAUUUUAAAAUUGAUUACCACGAGAAGGCAGUUUGGUAGUUUCCAUGUUGCAGUUACUCCAACUCCAACUCCG ..(((((((.....)))))))......(((((((...((((((....((((........))))...))))))..)))))))((......))..((((.((((.....)))).)))).... ( -26.70) >DroSim_CAF1 19959 120 - 1 ACAUGCCGUGCUUAAUGGCUUAAGUUAAAGCUGCAAAUUUUGUCAUAAAUUUUAAAAUUGAUUACCACGAGAAGGCAGUUUGGUAGUUUCCAUGUUGCAGUUACUCCAACUCCAACUCCG ....(((((.....)))))........(((((((...((((((....((((........))))...))))))..)))))))((......))..((((.((((.....)))).)))).... ( -24.20) >DroEre_CAF1 18936 117 - 1 ACAUGCCGUGCUUAAUGGCAUAAUUUAAAGCUGCAAAUUUUGUCAUAAAUUUUAAAAUUGAUUACCACGAGAAGGCAGUUUGGUAGUUUCCAUGUUGCAGUUACUCCAACUCCGACU--- ..(((((((.....)))))))......(((((((...((((((....((((........))))...))))))..)))))))((......))..((((.((((.....)))).)))).--- ( -26.40) >DroYak_CAF1 19368 117 - 1 ACAUGCCGUGCUUAAUGGCAUAAUUUAAAGCUGCAAAUUUUGUCAUAAAUUUUAAAAUUGAUUACCACGAGAAGGCAGUUUGGUAGUUUCCAUGUUGCAGUUACUCCAACUCCAACU--- ..(((((((.....)))))))......(((((((...((((((....((((........))))...))))))..)))))))((......))..((((.((((.....)))).)))).--- ( -26.70) >consensus ACAUGCCGUGCUUAAUGGCAUAAUUUAAAGCUGCAAAUUUUGUCAUAAAUUUUAAAAUUGAUUACCACGAGAAGGCAGUUUGGUAGUUUCCAUGUUGCAGUUACUCCAACUCCAACUCCG ..(((((((.....)))))))......(((((((...((((((....((((........))))...))))))..)))))))((......))..((((.((((.....)))).)))).... (-26.12 = -26.00 + -0.12)

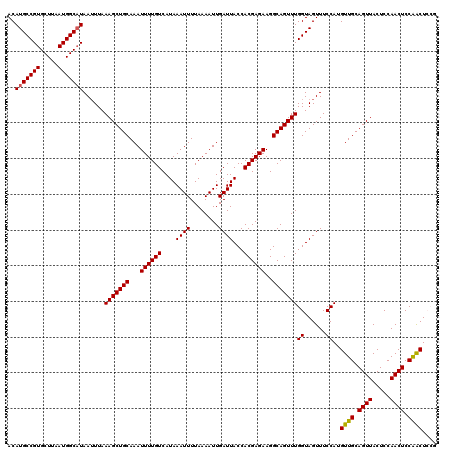
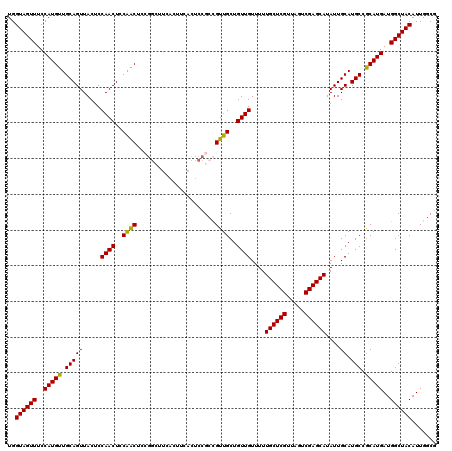
| Location | 14,369,848 – 14,369,968 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 99.33 |
| Mean single sequence MFE | -34.58 |
| Consensus MFE | -33.88 |
| Energy contribution | -34.28 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.98 |
| SVM decision value | 3.03 |
| SVM RNA-class probability | 0.998175 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14369848 120 - 22407834 GCAGUCGGGCUAACAGGCAGGCAGCCUGACGAUAUGAGUUACAUGCCGUGCUUAAUGGCAUAAUUUAAAGCUGCAAAUUUUGUCAUAAAUUUUAAAAUUGAUUACCACGAGAAGGCAGUU ...((((((((..(.....)..))))))))....(((((((..((((((.....))))))))))))).((((((...((((((....((((........))))...))))))..)))))) ( -35.10) >DroSec_CAF1 17898 120 - 1 GCAGUCGGGCUAACAGGCAGGCAGCCUGACGAUAUGAGUUACAUGCCGUGCUUAAUGGCAUAAUUUAAAGCUGCAAAUUUUGUCAUAAAUUUUAAAAUUGAUUACCACGAGAAGGCAGUU ...((((((((..(.....)..))))))))....(((((((..((((((.....))))))))))))).((((((...((((((....((((........))))...))))))..)))))) ( -35.10) >DroSim_CAF1 19999 120 - 1 GCAGUCGGGCUAACAGGCAGGCAGCCUGACGAUAUGAGUUACAUGCCGUGCUUAAUGGCUUAAGUUAAAGCUGCAAAUUUUGUCAUAAAUUUUAAAAUUGAUUACCACGAGAAGGCAGUU ...((((((((..(.....)..))))))))....(((.(((...(((((.....))))).))).))).((((((...((((((....((((........))))...))))))..)))))) ( -32.50) >DroEre_CAF1 18973 120 - 1 GCAGUCGGGCUAACAGGCAGGCAGCCUGACGAUAUGAGUUACAUGCCGUGCUUAAUGGCAUAAUUUAAAGCUGCAAAUUUUGUCAUAAAUUUUAAAAUUGAUUACCACGAGAAGGCAGUU ...((((((((..(.....)..))))))))....(((((((..((((((.....))))))))))))).((((((...((((((....((((........))))...))))))..)))))) ( -35.10) >DroYak_CAF1 19405 120 - 1 GCAGUCGGGCUAACAGGCAGGCAGCCUGACGAUAUGAGUUACAUGCCGUGCUUAAUGGCAUAAUUUAAAGCUGCAAAUUUUGUCAUAAAUUUUAAAAUUGAUUACCACGAGAAGGCAGUU ...((((((((..(.....)..))))))))....(((((((..((((((.....))))))))))))).((((((...((((((....((((........))))...))))))..)))))) ( -35.10) >consensus GCAGUCGGGCUAACAGGCAGGCAGCCUGACGAUAUGAGUUACAUGCCGUGCUUAAUGGCAUAAUUUAAAGCUGCAAAUUUUGUCAUAAAUUUUAAAAUUGAUUACCACGAGAAGGCAGUU ...((((((((..(.....)..))))))))....(((((((..((((((.....))))))))))))).((((((...((((((....((((........))))...))))))..)))))) (-33.88 = -34.28 + 0.40)

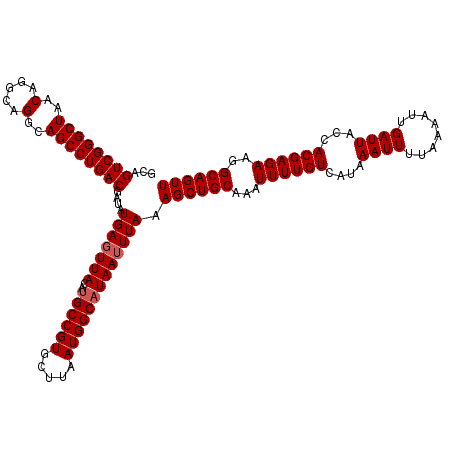
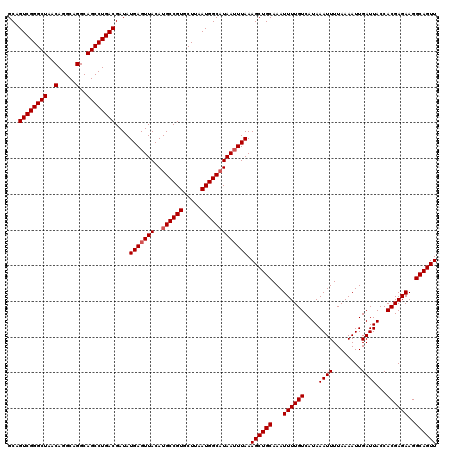
| Location | 14,369,888 – 14,370,008 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.50 |
| Mean single sequence MFE | -33.68 |
| Consensus MFE | -33.68 |
| Energy contribution | -33.68 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.23 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.729088 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14369888 120 + 22407834 AAAUUUGCAGCUUUAAAUUAUGCCAUUAAGCACGGCAUGUAACUCAUAUCGUCAGGCUGCCUGCCUGUUAGCCCGACUGCCUGGUGGCGGCUUGAAACAAGAAAAAGCCGAAGGAGAAAC ..........((((.......(((((((.(((.(..(((.....)))..)(((.(((((.(.....).))))).)))))).)))))))(((((...........)))))))))....... ( -34.00) >DroSec_CAF1 17938 120 + 1 AAAUUUGCAGCUUUAAAUUAUGCCAUUAAGCACGGCAUGUAACUCAUAUCGUCAGGCUGCCUGCCUGUUAGCCCGACUGCCUGGUGGCGGCUUGAAACAAGAAAAAGCCGAAGGAGAAAC ..........((((.......(((((((.(((.(..(((.....)))..)(((.(((((.(.....).))))).)))))).)))))))(((((...........)))))))))....... ( -34.00) >DroSim_CAF1 20039 120 + 1 AAAUUUGCAGCUUUAACUUAAGCCAUUAAGCACGGCAUGUAACUCAUAUCGUCAGGCUGCCUGCCUGUUAGCCCGACUGCAUGGUGGCGGCUUGAAACAAGAAAAAGCCGAAGGAGAAAC ..........((((.......(((((((.(((.(..(((.....)))..)(((.(((((.(.....).))))).)))))).)))))))(((((...........)))))))))....... ( -34.20) >DroEre_CAF1 19013 120 + 1 AAAUUUGCAGCUUUAAAUUAUGCCAUUAAGCACGGCAUGUAACUCAUAUCGUCAGGCUGCCUGCCUGUUAGCCCGACUGCCUGGUGGCGGCUUAAAACAAGAAAAAGCCGAAGGAGAAAC ..........((((.......(((((((.(((.(..(((.....)))..)(((.(((((.(.....).))))).)))))).)))))))(((((...........)))))))))....... ( -33.10) >DroYak_CAF1 19445 120 + 1 AAAUUUGCAGCUUUAAAUUAUGCCAUUAAGCACGGCAUGUAACUCAUAUCGUCAGGCUGCCUGCCUGUUAGCCCGACUGCCUGGUGGCGGCUUAAAACAAGAAAAAGCCGAAGGAGAAAC ..........((((.......(((((((.(((.(..(((.....)))..)(((.(((((.(.....).))))).)))))).)))))))(((((...........)))))))))....... ( -33.10) >consensus AAAUUUGCAGCUUUAAAUUAUGCCAUUAAGCACGGCAUGUAACUCAUAUCGUCAGGCUGCCUGCCUGUUAGCCCGACUGCCUGGUGGCGGCUUGAAACAAGAAAAAGCCGAAGGAGAAAC ..........((((.......(((((((.(((.(..(((.....)))..)(((.(((((.(.....).))))).)))))).)))))))(((((...........)))))))))....... (-33.68 = -33.68 + -0.00)

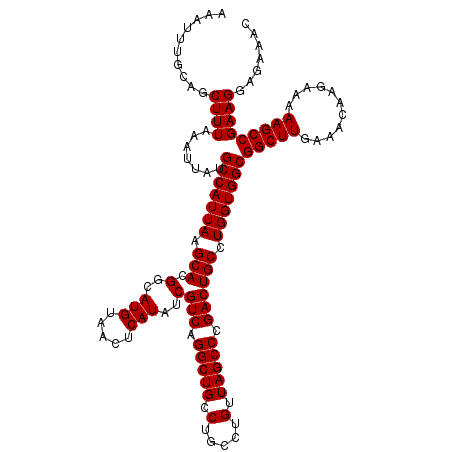
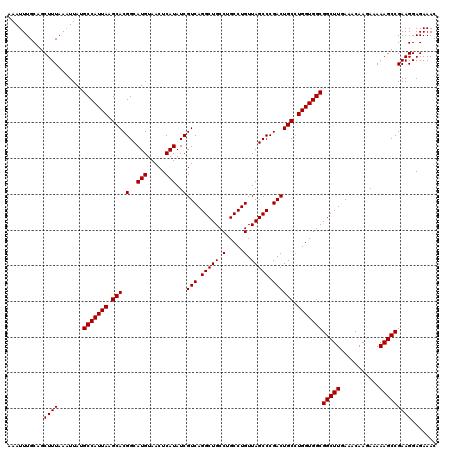
| Location | 14,369,888 – 14,370,008 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.50 |
| Mean single sequence MFE | -38.18 |
| Consensus MFE | -36.88 |
| Energy contribution | -37.28 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.97 |
| SVM decision value | 2.87 |
| SVM RNA-class probability | 0.997497 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14369888 120 - 22407834 GUUUCUCCUUCGGCUUUUUCUUGUUUCAAGCCGCCACCAGGCAGUCGGGCUAACAGGCAGGCAGCCUGACGAUAUGAGUUACAUGCCGUGCUUAAUGGCAUAAUUUAAAGCUGCAAAUUU ((..(((..(((((((...........)))))(((....))).((((((((..(.....)..))))))))))...)))..))(((((((.....)))))))................... ( -38.60) >DroSec_CAF1 17938 120 - 1 GUUUCUCCUUCGGCUUUUUCUUGUUUCAAGCCGCCACCAGGCAGUCGGGCUAACAGGCAGGCAGCCUGACGAUAUGAGUUACAUGCCGUGCUUAAUGGCAUAAUUUAAAGCUGCAAAUUU ((..(((..(((((((...........)))))(((....))).((((((((..(.....)..))))))))))...)))..))(((((((.....)))))))................... ( -38.60) >DroSim_CAF1 20039 120 - 1 GUUUCUCCUUCGGCUUUUUCUUGUUUCAAGCCGCCACCAUGCAGUCGGGCUAACAGGCAGGCAGCCUGACGAUAUGAGUUACAUGCCGUGCUUAAUGGCUUAAGUUAAAGCUGCAAAUUU ..........(((((((..((((.........(..((((((..((((((((..(.....)..))))))))..)))).))..)..(((((.....))))).))))..)))))))....... ( -35.10) >DroEre_CAF1 19013 120 - 1 GUUUCUCCUUCGGCUUUUUCUUGUUUUAAGCCGCCACCAGGCAGUCGGGCUAACAGGCAGGCAGCCUGACGAUAUGAGUUACAUGCCGUGCUUAAUGGCAUAAUUUAAAGCUGCAAAUUU ((..(((..(((((((...........)))))(((....))).((((((((..(.....)..))))))))))...)))..))(((((((.....)))))))................... ( -39.30) >DroYak_CAF1 19445 120 - 1 GUUUCUCCUUCGGCUUUUUCUUGUUUUAAGCCGCCACCAGGCAGUCGGGCUAACAGGCAGGCAGCCUGACGAUAUGAGUUACAUGCCGUGCUUAAUGGCAUAAUUUAAAGCUGCAAAUUU ((..(((..(((((((...........)))))(((....))).((((((((..(.....)..))))))))))...)))..))(((((((.....)))))))................... ( -39.30) >consensus GUUUCUCCUUCGGCUUUUUCUUGUUUCAAGCCGCCACCAGGCAGUCGGGCUAACAGGCAGGCAGCCUGACGAUAUGAGUUACAUGCCGUGCUUAAUGGCAUAAUUUAAAGCUGCAAAUUU ((..(((..(((((((...........)))))(((....))).((((((((..(.....)..))))))))))...)))..))(((((((.....)))))))................... (-36.88 = -37.28 + 0.40)

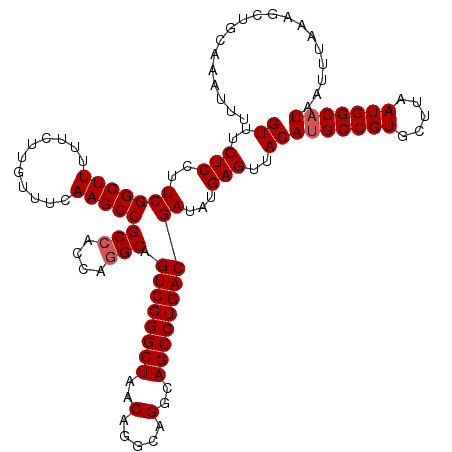
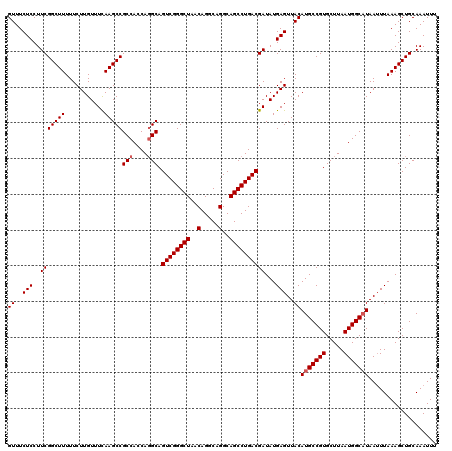
| Location | 14,369,928 – 14,370,048 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.67 |
| Mean single sequence MFE | -44.14 |
| Consensus MFE | -42.10 |
| Energy contribution | -42.54 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.95 |
| SVM decision value | 3.87 |
| SVM RNA-class probability | 0.999671 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14369928 120 + 22407834 AACUCAUAUCGUCAGGCUGCCUGCCUGUUAGCCCGACUGCCUGGUGGCGGCUUGAAACAAGAAAAAGCCGAAGGAGAAACAUGUUAACUGCUUUCCUUCGCCCCAGUGUGAGUACACCGG .(((((((..(((.(((((.(.....).))))).)))...((((.((((((((...........)))))((((((((..............))))))))))))))))))))))....... ( -45.34) >DroSec_CAF1 17978 120 + 1 AACUCAUAUCGUCAGGCUGCCUGCCUGUUAGCCCGACUGCCUGGUGGCGGCUUGAAACAAGAAAAAGCCGAAGGAGAAACAUGUUAACUGCUUUCCUUCGCCCCAGUGUGGAUACACCGG ......(((((((.(((((.(.....).))))).))).((((((.((((((((...........)))))((((((((..............))))))))))))))).)).))))...... ( -41.64) >DroSim_CAF1 20079 120 + 1 AACUCAUAUCGUCAGGCUGCCUGCCUGUUAGCCCGACUGCAUGGUGGCGGCUUGAAACAAGAAAAAGCCGAAGGAGAAACAUGUUAACUGCUUUCCUUCGCCCCAGUGUGAGUACACCGG .((((((((.(((.(((((.(.....).))))).)))....(((.((((((((...........)))))((((((((..............))))))))))))))))))))))....... ( -43.94) >DroEre_CAF1 19053 120 + 1 AACUCAUAUCGUCAGGCUGCCUGCCUGUUAGCCCGACUGCCUGGUGGCGGCUUAAAACAAGAAAAAGCCGAAGGAGAAACAUGUUAACUGCUUUCCUUCGCCCCAGUGUGAGUGCACCGG .(((((((..(((.(((((.(.....).))))).)))...((((.((((((((...........)))))((((((((..............))))))))))))))))))))))....... ( -44.44) >DroYak_CAF1 19485 120 + 1 AACUCAUAUCGUCAGGCUGCCUGCCUGUUAGCCCGACUGCCUGGUGGCGGCUUAAAACAAGAAAAAGCCGAAGGAGAAACAUGUUAACUGCUUUCCUUCGCCCCAGUGCGAGUGCACCGG .((((.....(((.(((((.(.....).))))).))).((((((.((((((((...........)))))((((((((..............))))))))))))))).))))))....... ( -45.34) >consensus AACUCAUAUCGUCAGGCUGCCUGCCUGUUAGCCCGACUGCCUGGUGGCGGCUUGAAACAAGAAAAAGCCGAAGGAGAAACAUGUUAACUGCUUUCCUUCGCCCCAGUGUGAGUACACCGG .(((((((..(((.(((((.(.....).))))).)))...((((.((((((((...........)))))((((((((..............))))))))))))))))))))))....... (-42.10 = -42.54 + 0.44)

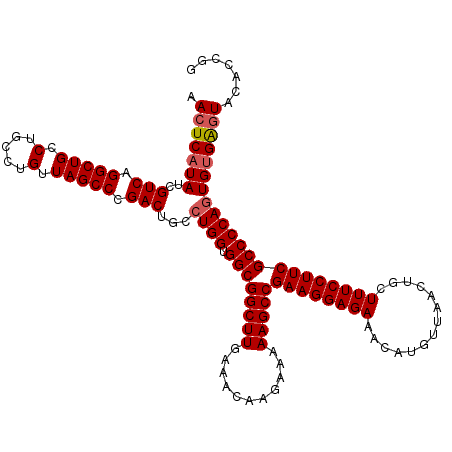
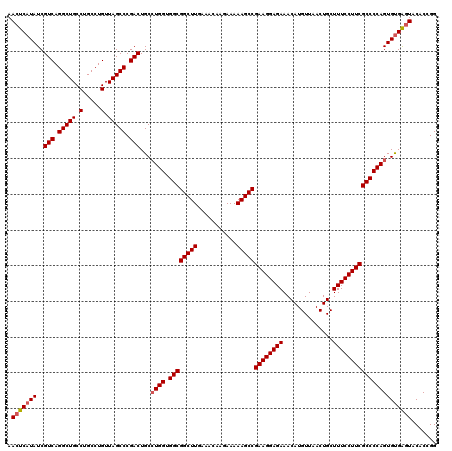
| Location | 14,369,928 – 14,370,048 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.67 |
| Mean single sequence MFE | -49.32 |
| Consensus MFE | -47.48 |
| Energy contribution | -47.44 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.27 |
| Structure conservation index | 0.96 |
| SVM decision value | 4.70 |
| SVM RNA-class probability | 0.999940 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14369928 120 - 22407834 CCGGUGUACUCACACUGGGGCGAAGGAAAGCAGUUAACAUGUUUCUCCUUCGGCUUUUUCUUGUUUCAAGCCGCCACCAGGCAGUCGGGCUAACAGGCAGGCAGCCUGACGAUAUGAGUU .......(((((..((((((((((((((((((.......))))).))))))(((((...........)))))))).))))...((((((((..(.....)..))))))))....))))). ( -48.50) >DroSec_CAF1 17978 120 - 1 CCGGUGUAUCCACACUGGGGCGAAGGAAAGCAGUUAACAUGUUUCUCCUUCGGCUUUUUCUUGUUUCAAGCCGCCACCAGGCAGUCGGGCUAACAGGCAGGCAGCCUGACGAUAUGAGUU (((((((....)))))))(.((((((((((((.......))))).))))))).).....((..(.((..(((.......))).((((((((..(.....)..)))))))))).)..)).. ( -47.60) >DroSim_CAF1 20079 120 - 1 CCGGUGUACUCACACUGGGGCGAAGGAAAGCAGUUAACAUGUUUCUCCUUCGGCUUUUUCUUGUUUCAAGCCGCCACCAUGCAGUCGGGCUAACAGGCAGGCAGCCUGACGAUAUGAGUU (((((((....)))))))((((((((((((((.......))))).))))))(((((...........))))))))((((((..((((((((..(.....)..))))))))..)))).)). ( -49.50) >DroEre_CAF1 19053 120 - 1 CCGGUGCACUCACACUGGGGCGAAGGAAAGCAGUUAACAUGUUUCUCCUUCGGCUUUUUCUUGUUUUAAGCCGCCACCAGGCAGUCGGGCUAACAGGCAGGCAGCCUGACGAUAUGAGUU .......(((((..((((((((((((((((((.......))))).))))))(((((...........)))))))).))))...((((((((..(.....)..))))))))....))))). ( -49.30) >DroYak_CAF1 19485 120 - 1 CCGGUGCACUCGCACUGGGGCGAAGGAAAGCAGUUAACAUGUUUCUCCUUCGGCUUUUUCUUGUUUUAAGCCGCCACCAGGCAGUCGGGCUAACAGGCAGGCAGCCUGACGAUAUGAGUU .......((((((.((((((((((((((((((.......))))).))))))(((((...........)))))))).)))))).((((((((..(.....)..)))))))).....)))). ( -51.70) >consensus CCGGUGUACUCACACUGGGGCGAAGGAAAGCAGUUAACAUGUUUCUCCUUCGGCUUUUUCUUGUUUCAAGCCGCCACCAGGCAGUCGGGCUAACAGGCAGGCAGCCUGACGAUAUGAGUU (((((((....)))))))((((((((((((((.......))))).))))))(((((...........))))))))((((....((((((((..(.....)..))))))))....)).)). (-47.48 = -47.44 + -0.04)

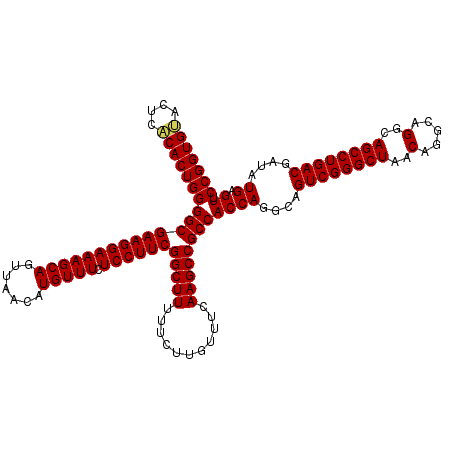
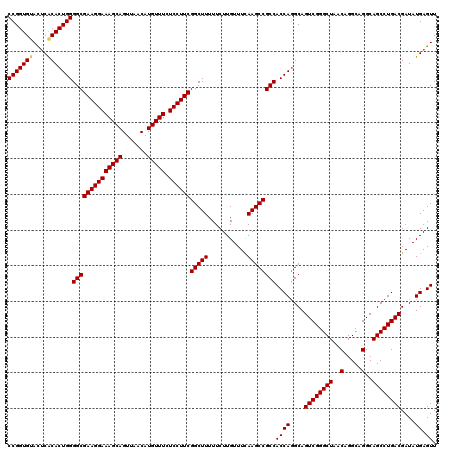
| Location | 14,369,968 – 14,370,075 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.17 |
| Mean single sequence MFE | -34.69 |
| Consensus MFE | -27.32 |
| Energy contribution | -28.44 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.79 |
| SVM decision value | -0.00 |
| SVM RNA-class probability | 0.532349 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14369968 107 + 22407834 CUGGUGGCGGCUUGAAACAAGAAAAAGCCGAAGGAGAAACAUGUUAACUGCUUUCCUUCGCCCCAGUGUGAGUACACCGGGAGAAAGGCAUAGUA-------------GCAACUUUAGUU ((((.((((((((...........)))))((((((((..............)))))))))))))))(((.(.((..((........))..)).).-------------)))......... ( -30.24) >DroSec_CAF1 18018 120 + 1 CUGGUGGCGGCUUGAAACAAGAAAAAGCCGAAGGAGAAACAUGUUAACUGCUUUCCUUCGCCCCAGUGUGGAUACACCGGGAGAAAUUCAUAGUAGGUCGUCAUAUAAGCCACUUUUCUU ((((.((((((((...........)))))((((((((..............))))))))))))))).((((..(((((.(((....)))......))).))........))))....... ( -34.34) >DroSim_CAF1 20119 120 + 1 AUGGUGGCGGCUUGAAACAAGAAAAAGCCGAAGGAGAAACAUGUUAACUGCUUUCCUUCGCCCCAGUGUGAGUACACCGGGAGAAAGUAAUAGUAGGUCAUCAUAUAAGCCACUUUUCUU ..(((((((((((...........)))))((((((((..............))))))))..(((.((((....)))).)))...........................))))))...... ( -33.64) >DroEre_CAF1 19093 110 + 1 CUGGUGGCGGCUUAAAACAAGAAAAAGCCGAAGGAGAAACAUGUUAACUGCUUUCCUUCGCCCCAGUGUGAGUGCACCGGGAGAAAAGCAUGGUCAGUGGUGCUGUAG----------AU ((((.((((((((...........)))))((((((((..............))))))))))))))).......((((((.((..(.....)..))..)))))).....----------.. ( -34.04) >DroYak_CAF1 19525 120 + 1 CUGGUGGCGGCUUAAAACAAGAAAAAGCCGAAGGAGAAACAUGUUAACUGCUUUCCUUCGCCCCAGUGCGAGUGCACCGGGAGAAAAGCGUGGCAGGUCGUGAUAUAUGCUCCUCUACUU ..(((((((((((...........)))))).(((((....((((((.(((((...(((..((((.((((....)))).))).)..)))...)))))....))))))...)))))))))). ( -41.20) >consensus CUGGUGGCGGCUUGAAACAAGAAAAAGCCGAAGGAGAAACAUGUUAACUGCUUUCCUUCGCCCCAGUGUGAGUACACCGGGAGAAAAGCAUAGUAGGUCGUCAUAUAAGCCACUUUACUU ..(((((((((((...........)))))((((((((..............))))))))..(((.((((....)))).)))...........................))))))...... (-27.32 = -28.44 + 1.12)


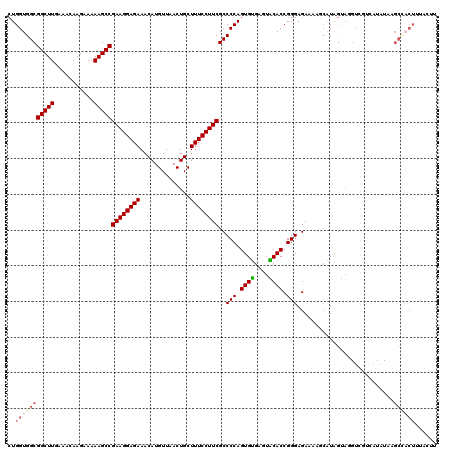
| Location | 14,369,968 – 14,370,075 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.17 |
| Mean single sequence MFE | -37.57 |
| Consensus MFE | -32.40 |
| Energy contribution | -32.36 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.12 |
| Structure conservation index | 0.86 |
| SVM decision value | 3.16 |
| SVM RNA-class probability | 0.998615 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14369968 107 - 22407834 AACUAAAGUUGC-------------UACUAUGCCUUUCUCCCGGUGUACUCACACUGGGGCGAAGGAAAGCAGUUAACAUGUUUCUCCUUCGGCUUUUUCUUGUUUCAAGCCGCCACCAG ....((((..((-------------......))))))...(((((((....)))))))((((((((((((((.......))))).))))))(((((...........))))))))..... ( -33.00) >DroSec_CAF1 18018 120 - 1 AAGAAAAGUGGCUUAUAUGACGACCUACUAUGAAUUUCUCCCGGUGUAUCCACACUGGGGCGAAGGAAAGCAGUUAACAUGUUUCUCCUUCGGCUUUUUCUUGUUUCAAGCCGCCACCAG .......(((((((....(((((...............(((((((((....)))))))))((((((((((((.......))))).)))))))........)))))..)))))))...... ( -38.90) >DroSim_CAF1 20119 120 - 1 AAGAAAAGUGGCUUAUAUGAUGACCUACUAUUACUUUCUCCCGGUGUACUCACACUGGGGCGAAGGAAAGCAGUUAACAUGUUUCUCCUUCGGCUUUUUCUUGUUUCAAGCCGCCACCAU .......(((((..........................(((((((((....))))))))).(((((((((((.......))))).))))))(((((...........))))))))))... ( -38.10) >DroEre_CAF1 19093 110 - 1 AU----------CUACAGCACCACUGACCAUGCUUUUCUCCCGGUGCACUCACACUGGGGCGAAGGAAAGCAGUUAACAUGUUUCUCCUUCGGCUUUUUCUUGUUUUAAGCCGCCACCAG ..----------.....(((((....................))))).......((((((((((((((((((.......))))).))))))(((((...........)))))))).)))) ( -36.15) >DroYak_CAF1 19525 120 - 1 AAGUAGAGGAGCAUAUAUCACGACCUGCCACGCUUUUCUCCCGGUGCACUCGCACUGGGGCGAAGGAAAGCAGUUAACAUGUUUCUCCUUCGGCUUUUUCUUGUUUUAAGCCGCCACCAG ....((((((((...................)))))))).(((((((....)))))))((((((((((((((.......))))).))))))(((((...........))))))))..... ( -41.71) >consensus AAGAAAAGUGGCUUAUAUCACGACCUACUAUGCCUUUCUCCCGGUGUACUCACACUGGGGCGAAGGAAAGCAGUUAACAUGUUUCUCCUUCGGCUUUUUCUUGUUUCAAGCCGCCACCAG ........................................(((((((....)))))))((((((((((((((.......))))).))))))(((((...........))))))))..... (-32.40 = -32.36 + -0.04)

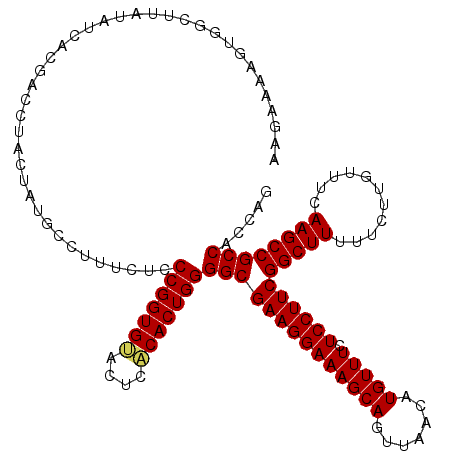
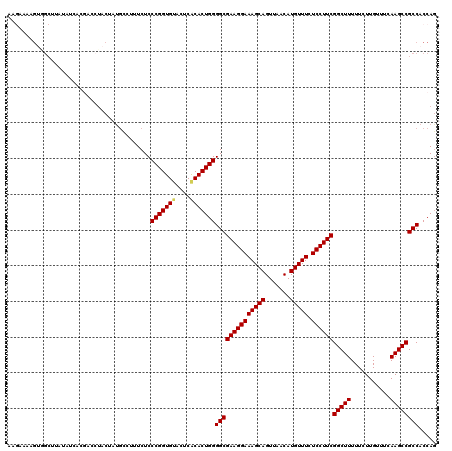
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:32:51 2006