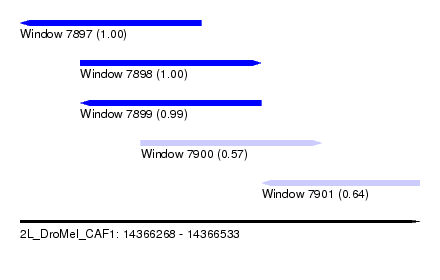
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,366,268 – 14,366,533 |
| Length | 265 |
| Max. P | 0.999744 |

| Location | 14,366,268 – 14,366,388 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 99.00 |
| Mean single sequence MFE | -39.14 |
| Consensus MFE | -38.64 |
| Energy contribution | -38.84 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.77 |
| Structure conservation index | 0.99 |
| SVM decision value | 3.99 |
| SVM RNA-class probability | 0.999744 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14366268 120 - 22407834 AAUCGUGGCAGACAAGCAUGCCGAACUGGCCGCAAAGGCAAUAUAUUUAAAGUUGCCUGUCAAUUUUGGAUGCUGUUAAGCGUUGCUAACACGCCAUGUUUGUUGAGUACGCGAUAUCAA .((((((((.((((((((((.((...((((.....(((((((.........)))))))..........((((((....))))))))))...)).))))))))))..)).))))))..... ( -39.40) >DroSec_CAF1 14239 120 - 1 AAUCGUGGCAGACAAGCAUGCCGAACUGGCCGCAAAGGCAAUAUAUUUAAAGUUGCCUGUCAAUUUUGGAUGCUGUUAAGCUUUGCUAACACGCCAUGUUUGUUGAGUACGCGAUAUCGA .((((((((.(((((((((((((((.((((.....(((((((.........)))))))))))..)))))..(((((((.((...))))))).))))))))))))..)).))))))..... ( -38.10) >DroSim_CAF1 16352 120 - 1 AAUCGUGGCAGACAAGCAUGCCGAACUGGCCGCAAAGGCAAUAUAUUUAAAGUUGCCUGUCAAUUUUGGAUGCUGUUAAGCGUUGCUAACACGCCAUGUUUGUUGAGUACGCGAUAUCAA .((((((((.((((((((((.((...((((.....(((((((.........)))))))..........((((((....))))))))))...)).))))))))))..)).))))))..... ( -39.40) >DroEre_CAF1 15191 120 - 1 AAUCGUGGCAGACAAGCAUGCCGAACUGGCCGCAAAGGCAAUAUAUUUAAAGUUGCCUGUCAAUUUUGGAUGCUGUUAAGCGUUGCUAACACGCCAUGUUUGUUGAGUACGCGAUAUCAA .((((((((.((((((((((.((...((((.....(((((((.........)))))))..........((((((....))))))))))...)).))))))))))..)).))))))..... ( -39.40) >DroYak_CAF1 14360 120 - 1 AAUCGUGGCAGACAAGCAUGCCGAACUGGCCGCAAAGGCAAUAUAUUUAAAGUUGCCUGUCAAUUUUGGAUGCUGUUAAGCGUUGCUAACACGCCAUGUUUGUUGAGUUCGCGAUAUCAA .((((((((.((((((((((.((...((((.....(((((((.........)))))))..........((((((....))))))))))...)).))))))))))..).)))))))..... ( -39.40) >consensus AAUCGUGGCAGACAAGCAUGCCGAACUGGCCGCAAAGGCAAUAUAUUUAAAGUUGCCUGUCAAUUUUGGAUGCUGUUAAGCGUUGCUAACACGCCAUGUUUGUUGAGUACGCGAUAUCAA .((((((((.((((((((((.((...((((.....(((((((.........)))))))..........((((((....))))))))))...)).))))))))))..)).))))))..... (-38.64 = -38.84 + 0.20)



| Location | 14,366,308 – 14,366,428 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.83 |
| Mean single sequence MFE | -38.88 |
| Consensus MFE | -38.22 |
| Energy contribution | -38.06 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.98 |
| SVM decision value | 2.56 |
| SVM RNA-class probability | 0.995268 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14366308 120 + 22407834 CUUAACAGCAUCCAAAAUUGACAGGCAACUUUAAAUAUAUUGCCUUUGCGGCCAGUUCGGCAUGCUUGUCUGCCACGAUUAGUGGCAAGCAGCCAGCCAGCGCGGCAAGCUCUGCGAUCC .......(((............((((((..(.....)..))))))..((.(((.(((.(((.((.(((..((((((.....))))))..))).))))))))..)))..))..)))..... ( -38.50) >DroSec_CAF1 14279 120 + 1 CUUAACAGCAUCCAAAAUUGACAGGCAACUUUAAAUAUAUUGCCUUUGCGGCCAGUUCGGCAUGCUUGUCUGCCACGAUUAGUGGCAAACAGCCAGCCAGCGCGGCAAGCUCUGCGAUCC .......(((............((((((..(.....)..))))))..((.(((.(((.(((.((.((((.((((((.....)))))).)))).))))))))..)))..))..)))..... ( -37.80) >DroSim_CAF1 16392 120 + 1 CUUAACAGCAUCCAAAAUUGACAGGCAACUUUAAAUAUAUUGCCUUUGCGGCCAGUUCGGCAUGCUUGUCUGCCACGAUUAGUGGCAAGCAGCCAGCCAGCGCGGCAAGCUCUGCGAUCC .......(((............((((((..(.....)..))))))..((.(((.(((.(((.((.(((..((((((.....))))))..))).))))))))..)))..))..)))..... ( -38.50) >DroEre_CAF1 15231 120 + 1 CUUAACAGCAUCCAAAAUUGACAGGCAACUUUAAAUAUAUUGCCUUUGCGGCCAGUUCGGCAUGCUUGUCUGCCACGAUUAGUGGCAAGCAGCCAGCCAGCGAGGCAAGCUCUGCGAUCC .......(((.......((((.((....)))))).....(((((((.(((((......(((.((((....((((((.....))))))))))))).))).)))))))))....)))..... ( -38.30) >DroYak_CAF1 14400 120 + 1 CUUAACAGCAUCCAAAAUUGACAGGCAACUUUAAAUAUAUUGCCUUUGCGGCCAGUUCGGCAUGCUUGUCUGCCACGAUUAGUGGCAAGCAGCCAGCCAGCGAGGCGAGCUCUGCGAUCC ......................((((((..(.....)..))))))((((((..((((((((.((((....((((((.....))))))))))))).(((.....))))))))))))))... ( -41.30) >consensus CUUAACAGCAUCCAAAAUUGACAGGCAACUUUAAAUAUAUUGCCUUUGCGGCCAGUUCGGCAUGCUUGUCUGCCACGAUUAGUGGCAAGCAGCCAGCCAGCGCGGCAAGCUCUGCGAUCC .......(((............((((((..(.....)..))))))..((.(((.(((.(((.((.((((.((((((.....)))))).)))).))))))))..)))..))..)))..... (-38.22 = -38.06 + -0.16)

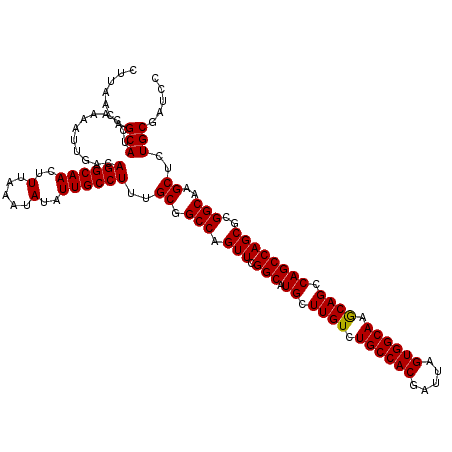

| Location | 14,366,308 – 14,366,428 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.83 |
| Mean single sequence MFE | -42.62 |
| Consensus MFE | -41.42 |
| Energy contribution | -41.26 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.97 |
| SVM decision value | 2.20 |
| SVM RNA-class probability | 0.990249 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14366308 120 - 22407834 GGAUCGCAGAGCUUGCCGCGCUGGCUGGCUGCUUGCCACUAAUCGUGGCAGACAAGCAUGCCGAACUGGCCGCAAAGGCAAUAUAUUUAAAGUUGCCUGUCAAUUUUGGAUGCUGUUAAG .....((((...(((((..((.(((((((((((((((((.....))))))....)))).))).....))))))...)))))..((((((((((((.....)))))))))))))))).... ( -42.10) >DroSec_CAF1 14279 120 - 1 GGAUCGCAGAGCUUGCCGCGCUGGCUGGCUGUUUGCCACUAAUCGUGGCAGACAAGCAUGCCGAACUGGCCGCAAAGGCAAUAUAUUUAAAGUUGCCUGUCAAUUUUGGAUGCUGUUAAG (.(((.((((((..((((...((((..((((((((((((.....))))))))).)))..))))...)))).))..(((((((.........)))))))......))))))).)....... ( -44.30) >DroSim_CAF1 16392 120 - 1 GGAUCGCAGAGCUUGCCGCGCUGGCUGGCUGCUUGCCACUAAUCGUGGCAGACAAGCAUGCCGAACUGGCCGCAAAGGCAAUAUAUUUAAAGUUGCCUGUCAAUUUUGGAUGCUGUUAAG .....((((...(((((..((.(((((((((((((((((.....))))))....)))).))).....))))))...)))))..((((((((((((.....)))))))))))))))).... ( -42.10) >DroEre_CAF1 15231 120 - 1 GGAUCGCAGAGCUUGCCUCGCUGGCUGGCUGCUUGCCACUAAUCGUGGCAGACAAGCAUGCCGAACUGGCCGCAAAGGCAAUAUAUUUAAAGUUGCCUGUCAAUUUUGGAUGCUGUUAAG .....((((...((((((.((.(((((((((((((((((.....))))))....)))).))).....))))))..))))))..((((((((((((.....)))))))))))))))).... ( -42.30) >DroYak_CAF1 14400 120 - 1 GGAUCGCAGAGCUCGCCUCGCUGGCUGGCUGCUUGCCACUAAUCGUGGCAGACAAGCAUGCCGAACUGGCCGCAAAGGCAAUAUAUUUAAAGUUGCCUGUCAAUUUUGGAUGCUGUUAAG .....(((..(((.(((.....))).)))))).((((((.....))))))((((.((((.(((((.((((.....(((((((.........)))))))))))..)))))))))))))... ( -42.30) >consensus GGAUCGCAGAGCUUGCCGCGCUGGCUGGCUGCUUGCCACUAAUCGUGGCAGACAAGCAUGCCGAACUGGCCGCAAAGGCAAUAUAUUUAAAGUUGCCUGUCAAUUUUGGAUGCUGUUAAG .....(((..(((.(((.....))).)))))).((((((.....))))))((((.((((.(((((.((((.....(((((((.........)))))))))))..)))))))))))))... (-41.42 = -41.26 + -0.16)


| Location | 14,366,348 – 14,366,468 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.18 |
| Mean single sequence MFE | -37.88 |
| Consensus MFE | -34.92 |
| Energy contribution | -35.12 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.574267 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14366348 120 + 22407834 UGCCUUUGCGGCCAGUUCGGCAUGCUUGUCUGCCACGAUUAGUGGCAAGCAGCCAGCCAGCGCGGCAAGCUCUGCGAUCCGGCUCCAAACCCAUAUCCAUCCCCAUGCCCAUCCCCAUGC ((..((((.((((.(.(((.((.(((((((((((((.....)))))).((.((......)))))))))))..))))).).)))).))))..))..........((((........)))). ( -38.30) >DroSec_CAF1 14319 104 + 1 UGCCUUUGCGGCCAGUUCGGCAUGCUUGUCUGCCACGAUUAGUGGCAAACAGCCAGCCAGCGCGGCAAGCUCUGCGAUCCGGCUUCAACCCCAGCCCCAUCAC------C---------- .((....((.(((.(((.(((.((.((((.((((((.....)))))).)))).))))))))..)))..))...))(((..((((........))))..)))..------.---------- ( -37.80) >DroSim_CAF1 16432 114 + 1 UGCCUUUGCGGCCAGUUCGGCAUGCUUGUCUGCCACGAUUAGUGGCAAGCAGCCAGCCAGCGCGGCAAGCUCUGCGAUCCGGCUUCAACCCCAUACCCAUCGC------CAUCACCAUGC .(((.((((((((.....)))..(((((((((((((.....)))))).((.((......)))))))))))..)))))...)))....................------........... ( -36.40) >DroEre_CAF1 15271 120 + 1 UGCCUUUGCGGCCAGUUCGGCAUGCUUGUCUGCCACGAUUAGUGGCAAGCAGCCAGCCAGCGAGGCAAGCUCUGCGAUCCGGCUCCAAUCCCAUCCCCAUCCCCAUACCCAUUCCCAGCC .(((.((((((..((((.(((.((((....((((((.....))))))))))))).(((.....))).))))))))))...)))..................................... ( -36.20) >DroYak_CAF1 14440 114 + 1 UGCCUUUGCGGCCAGUUCGGCAUGCUUGUCUGCCACGAUUAGUGGCAAGCAGCCAGCCAGCGAGGCGAGCUCUGCGAUCCGGCUCC------AUCCCCAUCUCCAUCCCCAUUCCCAUCC .(((.((((((..((((((((.((((....((((((.....))))))))))))).(((.....))))))))))))))...)))...------............................ ( -40.70) >consensus UGCCUUUGCGGCCAGUUCGGCAUGCUUGUCUGCCACGAUUAGUGGCAAGCAGCCAGCCAGCGCGGCAAGCUCUGCGAUCCGGCUCCAACCCCAUCCCCAUCCCCAU_CCCAUCCCCAUCC .(((.((((((..((((.(((.((((....((((((.....))))))))))))).(((.....))).))))))))))...)))..................................... (-34.92 = -35.12 + 0.20)



| Location | 14,366,428 – 14,366,533 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.00 |
| Mean single sequence MFE | -40.30 |
| Consensus MFE | -12.86 |
| Energy contribution | -14.26 |
| Covariance contribution | 1.40 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.32 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.641533 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14366428 105 - 22407834 CGUCUUCGCCCGAUUCACCCAAAUGAAGUUGGCUCUGAAGGAGC-UGAC-------UGUGUGG-AUGGCG-AUGG-----GCAUGGGGAUGGGCAUGGGGAUGGAUAUGGGUUUGGAGCC ((((((((((((..((.((((.....(((..((((.....))))-..))-------)......-...((.-....-----)).)))))))))))..)))))))......(((.....))) ( -36.30) >DroSec_CAF1 14399 87 - 1 CGUCUUCGCCCCAUUCACCCAAAUGAAGUUGGCUCUGAAGGAGC-UGGC-------UGUGUGG-AUGGCG-AU-----------------G------GUGAUGGGGCUGGGGUUGAAGCC ((.(((((((((((.((((.......(((..((((.....))))-..))-------).(((..-...)))-..-----------------)------)))))))))).)))).))..... ( -34.80) >DroSim_CAF1 16512 99 - 1 CGUCUUCGCCCCAUUCACCCAAAUGAAGUUGGCUCUGAAGGAGC-UGGC-------UGUGUGG-AUGGCG-AUGG-----GCAUGGUGAUG------GCGAUGGGUAUGGGGUUGAAGCC .(.(((((((((((..(((((.....(((..((((.....))))-..))-------).(((.(-.(.((.-((..-----..)).)).)).------))).))))))))))).)))))). ( -38.10) >DroEre_CAF1 15351 112 - 1 CGUCUUCGCCCCAUUCACCCAAAUGAAGUUGGCUCUGAAGGAGCCUGGC-------AGUGUGG-AUGGGCCACUGGAGUUGGCUGGGAAUGGGUAUGGGGAUGGGGAUGGGAUUGGAGCC ((((((((((((((..(((((.....(((..(((((..((..((((..(-------......)-..))))..)))))))..))).....))))))))))).))))))))........... ( -47.50) >DroYak_CAF1 14520 114 - 1 CGUCUUCGCCCCAUUCACCCAAAUGAAGUUGGCUCUGAAGGAGCCUCGCUGGCUGGAGUGUGGAAUGGGGCACGGGAGUUGGAUGGGAAUGGGGAUGGAGAUGGGGAU------GGAGCC ((((((((((((((((.((...........(((((.....)))))...(..(((...((((........))))...)))..)..)))))))))).)))))))).....------...... ( -44.80) >consensus CGUCUUCGCCCCAUUCACCCAAAUGAAGUUGGCUCUGAAGGAGC_UGGC_______UGUGUGG_AUGGCG_AUGG_____GCAUGGGAAUGGG_AUGGGGAUGGGGAUGGGGUUGGAGCC .(.((((..(((((...((((......((((((((.....)))).))))..........((......))................................)))).)))))...))))). (-12.86 = -14.26 + 1.40)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:32:33 2006