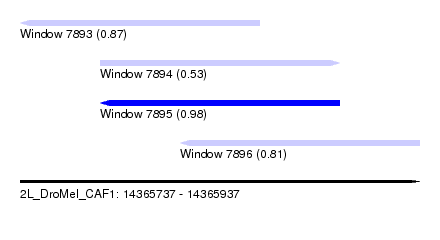
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,365,737 – 14,365,937 |
| Length | 200 |
| Max. P | 0.978231 |

| Location | 14,365,737 – 14,365,857 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.49 |
| Mean single sequence MFE | -35.78 |
| Consensus MFE | -29.73 |
| Energy contribution | -30.24 |
| Covariance contribution | 0.51 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.869890 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14365737 120 - 22407834 ACUGACGGCUGAGGUUAAAGCCCGGCCGAUUAAGCUGACAGUUUUUGUCGGGCUCGUAUUUUUCAGUUAAAAAGCGUGUUCAAUGUGACUUGGCACACCGCCGAAGAAAUACAUCGUGCU .....((((((.(((....)))))))))......(((((((...)))))))((.((....((((.........((((.....))))...(((((.....))))).)))).....)).)). ( -34.50) >DroSec_CAF1 13433 105 - 1 ACUGACGGCUGAGGUUAAAGCCCGGCCGAUUAAGCUGACAGUU--UGUCGGGCUCGUAUUUUUCAGUU---------UUUCAA----ACUUGGCACACAGGCGAAGAAAUACAUCGUGCU .(((.((((((.(((....))))))))).....((..(..(((--((..(((((.(......).))))---------)..)))----)))..))...)))((((.(.....).))))... ( -29.90) >DroSim_CAF1 15564 118 - 1 ACUGACGGCUGAGGUUAAAGCCCGGCCGAUUAAGCUGACAGUU--UGUCGGGCUCGUAUUUUUCAGUUAAAAAGCUUGUUCAAUGCGACUUGGCACACAGCCGAAGAAAUACAUCGUGCU .((..((((((.((((((.(((((((.((((........))))--.)))))))(((((((...(((((....))).))...))))))).))))).).)))))).)).............. ( -36.50) >DroEre_CAF1 14167 118 - 1 ACUGACGGCUGAGGUUAAAGCCCGGCCGAUUAAACUGACAGUU--UGUCGGGCUCGUAUUUUUCAGUUAAAAAGCGUGUUCAAUGUGACUUGGCACACAGCCGAAGAAAUACAUUGCGCU (((((((((((.(((....)))))))))......(((((....--.)))))...........))))).....((((....(((((((.((((((.....))).)))...))))))))))) ( -38.60) >DroYak_CAF1 13726 118 - 1 ACUGACGGCUGAGGUUAAAACCCGGCCGAUUAAACUGACAGUU--UGUCAGGCUCGUAUUUUUCAGUUAAAAAGCGUGUUCAAUGUGUCUUGGCACACAGCCGAACAAAUACAUAGCGCU (((((((((((.(((....)))))))))......(((((....--.)))))...........))))).....(((((........(((.(((((.....))))))))........))))) ( -39.39) >consensus ACUGACGGCUGAGGUUAAAGCCCGGCCGAUUAAGCUGACAGUU__UGUCGGGCUCGUAUUUUUCAGUUAAAAAGCGUGUUCAAUGUGACUUGGCACACAGCCGAAGAAAUACAUCGUGCU .....((((((.(((....)))))))))......((((((.....))))))....((((((((..........((((.....))))...(((((.....)))))))))))))........ (-29.73 = -30.24 + 0.51)

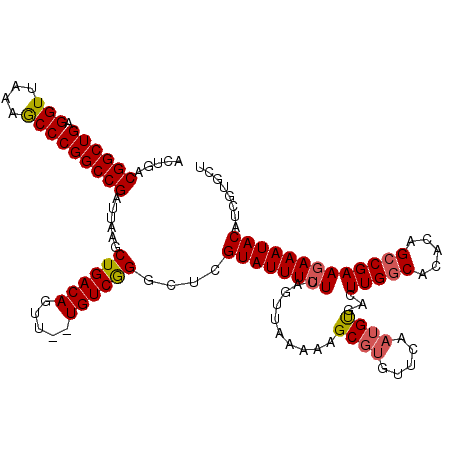
| Location | 14,365,777 – 14,365,897 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.26 |
| Mean single sequence MFE | -34.85 |
| Consensus MFE | -30.12 |
| Energy contribution | -30.32 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.86 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.528845 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14365777 120 + 22407834 AACACGCUUUUUAACUGAAAAAUACGAGCCCGACAAAAACUGUCAGCUUAAUCGGCCGGGCUUUAACCUCAGCCGUCAGUUGGUCUAAAGCGCCGCCGUUUCGCCAGCCGAUCUUUAGCG ....((((......((((.......((((((((((.....)))).(((.....))).)))))).....))))..(((.((((((...(((((....))))).)))))).)))....)))) ( -33.10) >DroSec_CAF1 13469 109 + 1 AAA---------AACUGAAAAAUACGAGCCCGACA--AACUGUCAGCUUAAUCGGCCGGGCUUUAACCUCAGCCGUCAGUUGGUCCAAAGCGCCGCCGUUUCGCCGGCCGAUCUUUAGCG ...---------..(((((......((((..(((.--....))).)))).((((((((((((((....(((((.....)))))...)))))..((......))))))))))).))))).. ( -32.00) >DroSim_CAF1 15604 118 + 1 AACAAGCUUUUUAACUGAAAAAUACGAGCCCGACA--AACUGUCAGCUUAAUCGGCCGGGCUUUAACCUCAGCCGUCAGUUGGUCCAAAGCGCCGCCGUUUCGCAGGCCGAUCUUUAGCG .....(((.................((((..(((.--....))).)))).((((((((((((((....(((((.....)))))...))))).))((......)).)))))))....))). ( -33.10) >DroEre_CAF1 14207 118 + 1 AACACGCUUUUUAACUGAAAAAUACGAGCCCGACA--AACUGUCAGUUUAAUCGGCCGGGCUUUAACCUCAGCCGUCAGUUGGGCUAAAGCGCCGCCGCUUCGCCGGCCGAUCUUUAGCG ....((((....((((((......((....))...--.....))))))..(((((((((((((((.((.((((.....)))))).))))))((....))....)))))))))....)))) ( -39.14) >DroYak_CAF1 13766 118 + 1 AACACGCUUUUUAACUGAAAAAUACGAGCCUGACA--AACUGUCAGUUUAAUCGGCCGGGUUUUAACCUCAGCCGUCAGUUGGGCUAAAGCGCCGCCGUUUCGCUGGCCGAUCUUUAGCG ..........((((((((.......((((.((((.--....))))))))...((((.((((....))).).))))))))))))(((((((..(.((((......)))).)..))))))). ( -36.90) >consensus AACACGCUUUUUAACUGAAAAAUACGAGCCCGACA__AACUGUCAGCUUAAUCGGCCGGGCUUUAACCUCAGCCGUCAGUUGGUCUAAAGCGCCGCCGUUUCGCCGGCCGAUCUUUAGCG ..............(((((......((((..((((.....)))).)))).(((((((((((((((...(((((.....)))))..)))))).))((......)).))))))).))))).. (-30.12 = -30.32 + 0.20)

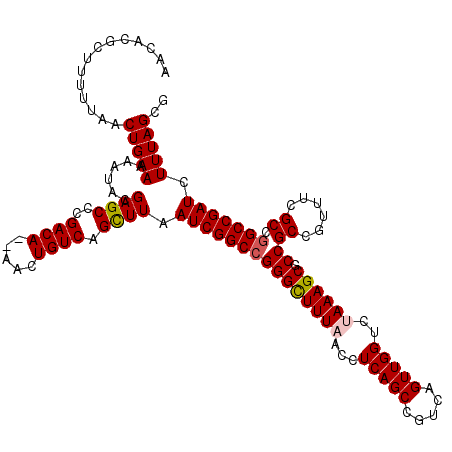
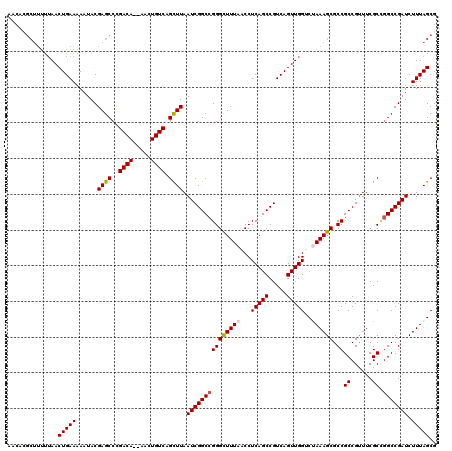
| Location | 14,365,777 – 14,365,897 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.26 |
| Mean single sequence MFE | -40.72 |
| Consensus MFE | -38.92 |
| Energy contribution | -38.60 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.81 |
| SVM RNA-class probability | 0.978231 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14365777 120 - 22407834 CGCUAAAGAUCGGCUGGCGAAACGGCGGCGCUUUAGACCAACUGACGGCUGAGGUUAAAGCCCGGCCGAUUAAGCUGACAGUUUUUGUCGGGCUCGUAUUUUUCAGUUAAAAAGCGUGUU ((((((((..(.((((......)))).)..)))))....((((((((((((.(((....)))))))))....(((((((((...))))).))))........)))))).....))).... ( -39.70) >DroSec_CAF1 13469 109 - 1 CGCUAAAGAUCGGCCGGCGAAACGGCGGCGCUUUGGACCAACUGACGGCUGAGGUUAAAGCCCGGCCGAUUAAGCUGACAGUU--UGUCGGGCUCGUAUUUUUCAGUU---------UUU ..((((((..(.((((......)))).)..))))))...((((((((((((.(((....)))))))))....(((((((....--.))).))))........))))))---------... ( -40.50) >DroSim_CAF1 15604 118 - 1 CGCUAAAGAUCGGCCUGCGAAACGGCGGCGCUUUGGACCAACUGACGGCUGAGGUUAAAGCCCGGCCGAUUAAGCUGACAGUU--UGUCGGGCUCGUAUUUUUCAGUUAAAAAGCUUGUU .((((((((((((((((((((....((((...(..(.....)..)((((((.(((....))))))))).....))))....))--)).))))).)).)))))).)))............. ( -37.80) >DroEre_CAF1 14207 118 - 1 CGCUAAAGAUCGGCCGGCGAAGCGGCGGCGCUUUAGCCCAACUGACGGCUGAGGUUAAAGCCCGGCCGAUUAAACUGACAGUU--UGUCGGGCUCGUAUUUUUCAGUUAAAAAGCGUGUU ((((...(((((((((((......))((((((((((((........)))))))))....))))))))))))...(((((....--.))))).....................)))).... ( -44.20) >DroYak_CAF1 13766 118 - 1 CGCUAAAGAUCGGCCAGCGAAACGGCGGCGCUUUAGCCCAACUGACGGCUGAGGUUAAAACCCGGCCGAUUAAACUGACAGUU--UGUCAGGCUCGUAUUUUUCAGUUAAAAAGCGUGUU .(((((((..(.(((........))).)..)))))))..((((((((((((.(((....)))))))))......(((((....--.)))))...........))))))............ ( -41.40) >consensus CGCUAAAGAUCGGCCGGCGAAACGGCGGCGCUUUAGACCAACUGACGGCUGAGGUUAAAGCCCGGCCGAUUAAGCUGACAGUU__UGUCGGGCUCGUAUUUUUCAGUUAAAAAGCGUGUU ..((((((..(.((((......)))).)..))))))...((((((((((((.(((....)))))))))......((((((.....))))))...........))))))............ (-38.92 = -38.60 + -0.32)


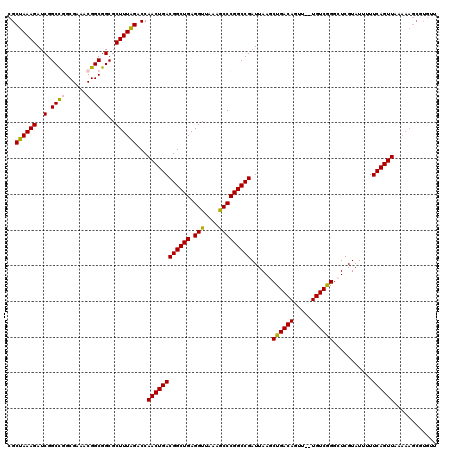
| Location | 14,365,817 – 14,365,937 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.92 |
| Mean single sequence MFE | -39.48 |
| Consensus MFE | -36.88 |
| Energy contribution | -36.48 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.814942 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14365817 120 - 22407834 CUUUUGACAGCACUUAAGUUGAAUUUGGUAAUGACAGUCGCGCUAAAGAUCGGCUGGCGAAACGGCGGCGCUUUAGACCAACUGACGGCUGAGGUUAAAGCCCGGCCGAUUAAGCUGACA .......((((.......................((((....((((((..(.((((......)))).)..))))))....)))).((((((.(((....))))))))).....))))... ( -38.80) >DroSec_CAF1 13498 120 - 1 CUUUUGACAGCACUUAAGUUGAAUUUGGUAAUGACAGUCGCGCUAAAGAUCGGCCGGCGAAACGGCGGCGCUUUGGACCAACUGACGGCUGAGGUUAAAGCCCGGCCGAUUAAGCUGACA .......((((.......................((((....((((((..(.((((......)))).)..))))))....)))).((((((.(((....))))))))).....))))... ( -40.70) >DroSim_CAF1 15642 120 - 1 CUUUUGACAGCACUUAAGUUGAAUUUGGUAAUGACAGUCGCGCUAAAGAUCGGCCUGCGAAACGGCGGCGCUUUGGACCAACUGACGGCUGAGGUUAAAGCCCGGCCGAUUAAGCUGACA .......((((.......................((((....((((((..(.(((........))).)..))))))....)))).((((((.(((....))))))))).....))))... ( -37.10) >DroEre_CAF1 14245 120 - 1 CUUUUGACAGCACUUAAGUUGAAUUUGGUAAUGACAGUCGCGCUAAAGAUCGGCCGGCGAAGCGGCGGCGCUUUAGCCCAACUGACGGCUGAGGUUAAAGCCCGGCCGAUUAAACUGACA .......((((......))))...(..((.....((((.(.(((((((....((((.((...)).)))).))))))).).)))).((((((.(((....))))))))).....))..).. ( -42.00) >DroYak_CAF1 13804 120 - 1 CUUUUGACAGCACUUAAGUUGAAUUUGGUAAUGACAGUCGCGCUAAAGAUCGGCCAGCGAAACGGCGGCGCUUUAGCCCAACUGACGGCUGAGGUUAAAACCCGGCCGAUUAAACUGACA .......((((......))))...(..((.....((((.(.(((((((..(.(((........))).)..))))))).).)))).((((((.(((....))))))))).....))..).. ( -38.80) >consensus CUUUUGACAGCACUUAAGUUGAAUUUGGUAAUGACAGUCGCGCUAAAGAUCGGCCGGCGAAACGGCGGCGCUUUAGACCAACUGACGGCUGAGGUUAAAGCCCGGCCGAUUAAGCUGACA .......((((......))))...(..((.....((((.(.(((((((..(.((((......)))).)..)))))).)).)))).((((((.(((....))))))))).....))..).. (-36.88 = -36.48 + -0.40)

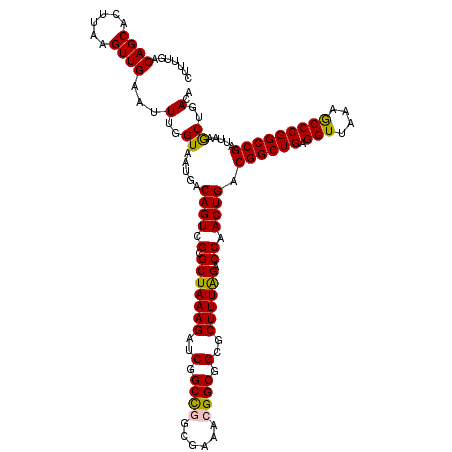

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:32:28 2006