
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,337,147 – 14,337,252 |
| Length | 105 |
| Max. P | 0.611372 |

| Location | 14,337,147 – 14,337,252 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 67.78 |
| Mean single sequence MFE | -29.60 |
| Consensus MFE | -8.40 |
| Energy contribution | -8.60 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.28 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.611372 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14337147 105 - 22407834 CCGGGGCUAUUCAAAGAUUUUUUUU------UACUCCCAAAUCCACUAGAAGGCAUCCUCCACUUGCCGA-GGCAUCCUGGA-GAUGCUCUAAG-----GAUAUGUAGACCCCUUAUU-- ..((((((((..(((((....))))------)........((((..((((..(((((.((((..(((...-.)))...))))-))))))))).)-----)))..)))).)))).....-- ( -32.40) >DroSec_CAF1 2925 103 - 1 CCGGGGGCAUUCAAAGAUUUUUUUU-----GUGCUUCCAAAUCCACGAGAAGGCAUCCU---CUUGCCGA-GGUAUCCUGGA-GAUGCUCUUAG-----GAUAUGUAGACCCCUUAUU-- ..((((((.(((........(((((-----(((..........))))))))((((....---..))))))-)((((((((((-(...))).)))-----)))))...).)))))....-- ( -31.00) >DroSim_CAF1 2935 86 - 1 CCGGGGGCAUUCAAAGUUUUUUUUU-----GUGCUUCCAAAUCCACGAG--------------------A-GGCAUCCUGGA-GAUGCUCUUAG-----GAUAUGUAGACCCCUUAUU-- ..(((((((..(((((.....))))-----))))...((.((((..(((--------------------(-(.((((.....-))))))))).)-----))).))....)))).....-- ( -25.60) >DroEre_CAF1 2894 92 - 1 CCGGGGGCAUUCACAGAUUUUUU----------CAUCCAAAUCUACCAGCAGGC---------UCUUCGA-GGCAUCCUGGA-GUUGCUCCGAG-----GAUAUGCAGACCCCUUAUU-- ..((((((......((((((...----------.....))))))....(((...---------((((((.-.(((.......-..)))..))))-----))..))).).)))))....-- ( -27.70) >DroYak_CAF1 3746 92 - 1 CCGGGGGCAUUCACAGAUUUUUU----------AUUCCAAGUCUACUAGAGGGC---------UCUCCGA-GGCAUCCUGGA-GAUGCUCUAAG-----GAUAUGUAGCCUCCUUACU-- ..((((((......((((((...----------.....)))))).((..(((((---------(((((.(-((...))))))-)).)))))..)-----).......)))))).....-- ( -32.40) >DroAna_CAF1 3034 106 - 1 CCCGGGCCAUUCACAGAUUUGUUUUCGCCUGUACUGGCAAAUUGA-----GGAU---------GCUCUGAAGUCGUCCACGAGGAGGCUCCAAGACAAGUAUCUGUAUCUGUAUGCCUUG ...((((.((..((((((........(((......))).......-----((((---------(((((..((((.(((....)))))))...)))...))))))..)))))))))))).. ( -28.50) >consensus CCGGGGGCAUUCAAAGAUUUUUUUU______U_CUUCCAAAUCCACGAGAAGGC_________UCUCCGA_GGCAUCCUGGA_GAUGCUCUAAG_____GAUAUGUAGACCCCUUAUU__ ..((((.........(((((..................)))))............................((((((......))))))....................))))....... ( -8.40 = -8.60 + 0.20)

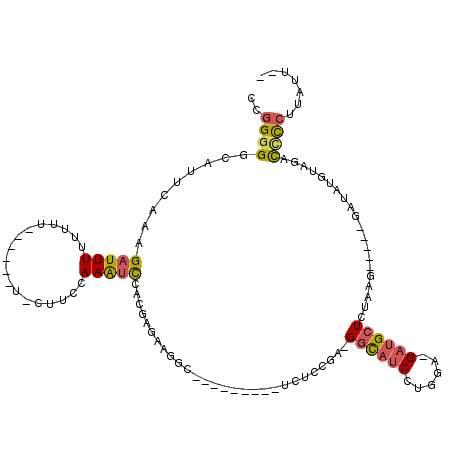
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:32:22 2006